Introduction
To assess the possibility that a foetus may be born with a genetic disorder or a pregnancy-related complication, prenatal screening is made available to all pregnant women. As a result, it assists in selecting various pregnancy alternatives or management strategies for the pregnancy and delivery in order to enhance the outcomes for the mother and the foetus [1]. According to the gestational trimester, in addition to the condition under consideration, there are many prenatal screening options [1]. A serious public health problem, foetal abnormalities are discovered in 2–3% of pregnancies [2–6], requiring further morphological testing, invasive procedures, genetic counselling to provide cytogenetics testing, and proper obstetric/perinatal care [7]. Following severe conditions, physical development abnormalities may appear, adding to the family’s mental and physical burden [8, 9]. Typically, a few ultrasonographic (US) studies are stated. The first-trimester sonographic scan evaluates the nasal bone presence and nuchal translucency (NT) while ruling out ductus venosus reversed flow and tricuspid regurgitation. In addition, the mid- trimester sonographic scan includes a systematic sequential examination of various foetal organs. The effectiveness of a sonographic scan for finding abnormalities ranges from 15 to 85%, based on the weeks of gestational age, foetal organ, sonographer’s proficiency, and body mass index of women [10]. Along with foetal structural defects or aberrant foetal development, several additional non-specific, frequently temporary anatomic findings, commonly referred to as “soft markers”, may also be found [11, 12]. Despite having no anatomical defects, these soft markers appear to be associated with a statistically higher risk of foetal aneuploidy [12–14].
Microarray analysis, commonly referred to as chromosomal microarray analysis (CMA) or molecular karyotyping, is slowly taking advantage of conventional G-banded karyotyping in order to the primary diagnostic test for adults and children who demonstrate a variety of neurodevelopmental phenotypes regardless of related congenital abnormalities in the past several years [15–18].
Invasive diagnostic testing generally is not suggested following the findings of a single soft marker or after low-risk screening assessments for aneuploidies – combined first-trimester screen including non-invasive prenatal testing (NIPT) or non-invasive prenatal screening (NIPS) for aneuploidies [12, 19, 20].
The soft markers were first identified as signs of aneuploidy, most notably Trisomy 21 (Down syndrome), as well as Trisomy 18 and Trisomy 13, which may be identified by each of the techniques mentioned above. Meanwhile, studies have indicated that certain soft markers can also be linked to submicroscopic chromosomal abnormalities that karyotyping or NIPT miss [21].
Chromosomal microarray analysis can be employed to find copy number variations (CNVs) within the genome, having a resolution of up to 10 kB. Additionally, the analysis might use single nucleotide polymorphism (SNP) array or else comparative genomic hybridisation (CGH) technology [22–24]. Sampling foetal cells now requires an invasive procedure, similar to karyotyping; however, the assay may be done on DNA isolated straight from foetal cells, avoiding the requirement for cell culture; consequently, findings are received faster [24]. Chromosomal microarray analysis has progressed into the first-tier test throughout the assessment of individuals with intellectual impairment, various congenital anomalies, and autism over the last decade [17]. The identification rate of possibly pathogenic CNVs (pCNVs) in foetuses with aberrant sonographic results varies from 5 to 8.5% in the prenatal setting, depending on the severity and type of congenital malformations [24]. As a result, CMA has replaced conventional karyotype as the primary suggested test in pregnancies presenting sonographic foetal anomalies in the structure [25]. Furthermore, evaluating variations of unknown significance (VOUS) in relation to the setting of an ongoing prenatal diagnosis is problematic [26, 27].
Ultrasonography is the primary clinical approach for identifying prenatal abnormalities at the moment [28], and it can identify structural abnormalities in the foetus as well as soft markers that suggest abnormalities [28]. Still, the diagnostic effectiveness of ultrasonography is limited [28, 29].
The limitations and difficulties of using only soft markers for anomaly detection will also be covered in this paper. We will discuss variables including operator skill, equipment quality, gestational age at assessment, maternal features, and potential confounding variables that may affect their accuracy. This study seeks to give healthcare practitioners a thorough understanding of the diagnostic efficacy of soft markers in determining abnormalities during pregnancy by critically reviewing the body of available literature. The results will improve prenatal counselling, direct clinical decision-making, and contribute to the creation of evidence-based procedures for the best prenatal care. Finally, for proper therapy and support throughout pregnancy, reliable foetal anomaly detection and assessment are essential. Regular ultrasound exams might uncover soft markers that can provide important information about probable anomalies. This systematic review aims to provide information on the diagnostic specificity of various markers, their relationship to particular anomalies, and the difficulties in interpreting them.
Material and methods
Search strategy
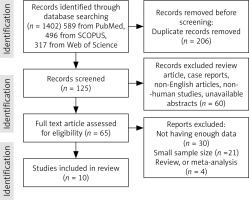
Electronic databases PubMed, Scopus, Google Scholar, Web of Sciences, and EMBASE were searched using the medical subject headings to identify all research articles related to the topic “The predictive and diagnostic value of ultrasound soft markers in the diagnostic of fetal abnormalities”. Two authors independently searched using search strategies specific for each database and reviewed all relevant peer-reviewed articles published before August 2023. The following search terms were used in our search strategy: (soft) and (biomarkers) or (biomarkers) or (marker) or (markers) and (microarray analysis) or (microarray) and (analysis) or (microarray analysis) or (microarray) or (microarrayed) or (microarray) or (microarrays). A total of 10 articles were included (Fig. 1).
Types of studies (selection criteria)
The included studies had to provide information on ultrasonic soft markers, foetal chromosomal abnormalities, pregnant women, and genetic tests like karyotyping, CMA, SNP array, and NIPT (or NIPS). Only articles published in English language were included. We excluded studies that evaluate specific syndromes related to specific chromosomal abnormalities and genes, and the ones that focused on molecules and tumours. Review articles, case reports, conference abstracts, case series, duplicate data, and low-quality studies were also excluded.
Data extraction
The relevant studies were selected according to the articles’ title, abstract, and full-text screening. In addition, the reference lists of selected studies were reviewed to identify any additional articles in case they were not recognised by the search process. The following information was extracted from each of the identified studies: author, year of publication, country, study design, total number of pregnant women, total number of foetuses, maternal mean age, gestational age, type of genetic tests, number of abnormal screening US soft markers, and types of abnormalities.
Limitations
The important variety in the study’s methodology, including changes in patient demographics, procedures, as well as study settings, and, is one important limitation seen throughout the investigated studies. Due to this diversity, it may be difficult to summarise the findings and make broad generalisations. The differences in the diagnostic precision predictions are introduced by the absence of established techniques for the evaluation of microarray analysis and ultrasound soft markers, potentially impacting the general validity of the results. Small sample sizes have been reported in some studies that were part of this systematic review, which may have an effect on the results statistical generalisability and reliability. Because of the small sample sizes, it is possible that they do not fully represent the prevalence of certain ultrasound markers in the overall population or the complete range of foetal abnormalities. So, when extending results to larger clinical practice, care should be taken. The knowledge and expertise of the sonographer applying the scan, in addition to the calibre of the ultrasound instruments employed, might have an impact on the precision of the ultrasound soft marker evaluation. The repeatability of outcomes in various clinical settings might be impacted by variations in operator skill and equipment capabilities that result in irregularities in marker detection and measurement. Variations of unknown significance or genetic abnormalities with unclear clinical use may be discovered by microarray analysis. Clinical decision-making might be complicated by the characterisation of VOUS because their importance may need to be determined by further investigation or long-term observation. The existence of VOUS makes advising pregnant parents more difficult and emphasises the continual requirement for genetic competence in result evaluation. Consideration should also be given to the accessibility microarray analysis and its affordability across diverse healthcare settings. To demonstrate significant relationships between microarray data and ultrasonic soft markers, future studies will need to emphasise large-scale, multicentre investigations that use standardised techniques. It is noteworthy that this study shows that the sensitivity and specificity in patients with 3 or more ultrasound soft markers are higher than the other groups but in comparison to karyotyping, CMA, SNP array, and NIPS, it still has lower sensitivity and specificity. Therefore it should still be considered that ultrasound soft markers alone are not yet fully reliable due to normal variations and the limitations mentioned above.
Result
Demographics and characteristics
Systematic review: Figure 1 summarises the studies chosen for the systematic review. Electronic and manual reference searches resulted in studies which were evaluated in full text and known were excluded (Table 1). To investigate the development of foetal abnormalities, such as foetal abnormalities and aneuploidies, 10 studies covering a total of 18,580 cases were included. These studies looked at sonographic markers, followed by a genetic diagnostic method called microarray analysis, which currently includes conventional karyotyping, SNP array, CMA, and NIPS, or NIPT, combined with imaging screening and diagnosing, incorporated into the review references, 9 of which included the collection of retrospective data, and one of cross-sectional data (Table 1). [30–33] of the 10 studies examined, utilised the CMA method to detect anomalies, 4 other distinct studies [34–37] used the SNP array approach, and one study [37] from the same group used conventional karyotyping. Finally, for the genetic analysis of the samples, one research [38] used the NIPS technique, and another [39] used the NIPT method. None the studies examined in this analysis had the required and accurate data for the groups we examined, including the average age of pregnant mothers and foetuses, and the calculation of the central tendency such as mean age for the entire study could not be done, but the values of some of them could be extracted from the studies and included of mean maternal ages in Cai et al. [37]: 28.9, Cai et al. [36]: 32.1 ±6.1 (18–46), Xiang et al. [34]: 32.04 (18–49), Beulen et al. [39]: 31 (17–44), Cai et al. [35]: (18–48), Bardin et al. [32]: 32 ±4.4, Ainsworth et al. [38]: 30.2 (8.2), Lostchuck et al. [33]: 32.2 (15–50), and Lin et al. [31]: were more than 24 years old, furthermore, mean gestational age (range) in Cai et al. [37]: 24.3 (13–38) weeks, Cai et al. [36]: 32.04 (18–49), Xiang et al. [34]: 21.28 (9–34), Beulen et al. [39]: 20, Cai et al. [35]: (18–48), Bardin et al. [32]: 32 ±4.4, Ainsworth et al. [38]: 30.2 (8.2), Lostchuck et al. [33]: 32.2 (15–50), and the range years of foetuses in Lin et al. [31] were 16–18 weeks.
Table 1
Overview of included studies
| Author | Year | Country/ continent | Study Design | Total number of pregnant women | Total number of fetuses | Maternal mean age (range) | Gestational age (mean) | Type of genetic tests | Number of abnormal screening ultrasonic soft markers | Types of abnormalities | Number of abnormal genetic tests within groups with abnormal soft markers in ultrasonography | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bornstein et al. [30] | 2016 | USA | Retros- pective | 1940 | 1940 | CMA | Ultrasonic soft marker anomalies, n = 359 of 536 (67%) | Pathologic n = 6, abnormal: CNV n = 4/359 (1.1%) – VOUS likely abnormal n = 2/359 (0.5%) | Prenatal CMA detected clinically relevant CNVs in fetuses with a normal karyotype | |||
| Beulen et al. [39] | 2016 | Europen | Retros- pective | 251 | 251 | 31 (17–44) | Mean = 20 | NIPT/diagnostic genetic test | Single marker, n = 45 of 73 (61.6%) | Trisomy 21 | NIPT n = 1 (1.3%)/diagnostic genetic test n = 1 (1.3%) | NIPT should not be recommended for the genetic evaluation of the aetiology of ultrasound anomalies, as both resolution and sensitivity, or negative predictive value, are inferior to those of conventional karyotyping and microarray analysis |
| Double markers , n = 22 of 73 (30.1%) | Trisomy 21 | NIPT n = 1 (1.3%)/diagnostic genetic test n = 1 (1.3%) | ||||||||||
| Multiple markers, n = 6 of 73 (8.2%) | Trisomy 21 | NIPT n = 0/diagnostic genetic test n = 0 | ||||||||||
| Cai et al. [37] | 2021 | Asia | Retros- pective | 1131 | 28.9 (18–47) | 13–38 weeks (24.3) | Conventional Karyotyping and SNP-array | Single ultrasonic soft marker, n = 729 | Trisomy 21 n = 12 of 729 (1.6%)/Trisomy18 n = 3 of 729 (0.4%)/Trisomy13 n = 3 of 729 (0.4%)/47, XXY n = 1 of 729 (0.13%)/45, X n = 2 of 729 (0.26%)/Chromosomal structural abnormalities n = 5 of 729 (0.68%)/abnormal CNVs n = 45 of 729 (6.1%) | Karyotype n = 26/SNP-array n = 45 | The SNP array can fully complement conventional karyotyping in fetuses with ultrasonic soft markers, improve detection rate of chromosomal abnormalities | |
| Two ultrasonic soft markers n = 322 | Trisomy 21, n = 7 of 322 (2.1%)/Trisomy 18 n = 1 of 322 (0.3%)/47, XXY n = 1 of 322 (0.3%)/mosaic chromosomal number abnormalities n = 1 of 322 (0.3%)/chromosomal structural abnormalities n = 3 of (0.9%)/abnormal CNV n = 20 of 322 (6.2%) | Karyotype n = 13/SNP-array n = 20 | ||||||||||
| Tree or more ultrasonic soft markers n = 80 | Trisomy 21 n = 4 of 80 (5%) /Trisomy 18 n = 1 of 80 (1.25%)/mosaic chromosomal number abnormalities n = 1 of 80 (1.25%)/chromosomal structural abnormalities n = 1 (1.25%)/abnormal CNV n = 4 of 80 (5%) | Karyotype n = 7/SNP-array n = 4 | ||||||||||
| Xiang et al. [34] | 2020 | Asia | Retros- pective | 5000 | 32.04 (18–49) | 9–34 weeks (21.28) | SNP-array | Single ultrasonographic soft marker n = 458 | Aneuploidy and Triploidy n = 24 of 458 (5.2%)/LOH n = 1 (0.2%)/CNV n = 30 of 458 (6.5%) | SNP n = 55 | SNP-array could additionally identify clinically significant submicroscopic CNVs, and we recommend the combination of SNP-array analysis and karyotyping in prenatal diagnosis | |
| Multiple ultrasonographic soft markers n = 72 | Aneuploidy and Triploidy n = 10 of 72 (13.8%)/CNV n = 5 of 72 (6.9%) | SNP n = 15 | ||||||||||
| Cai et al. [36] | 2020 | Asia | Retros- pective | 713 | 32.1 ±6.1 (18–46) | 28–37 (29.0 ±3.4) | SNP-array | n = 336 | CNV anomaly n = 16 of 336 (4.7%)/pCNVs n = 5 of 336 (1.4%)/VOUS n = 11 of 336 (3.2%) | SNP-array n = 16 | It is suggested that CMA should be considered for genetic analysis in cases with abnormal ultrasound fndings in the second and third trimesters of pregnancy | |
| Cai et al. [35] | 2023 | Asia | Retros- pective | 8386 | (18–48) | 11–36 weeks | SNP-array | Single ultrasonic soft marker n = 1949 | CNV anomaly: pCNV n = 89 of 1949 (4.6%)/VOUS n = 28 of 1949 1.4 (%) | SNP-array n = 117 | SNP-array should be performed on pregnant women when multiple ultrasound sof markers are detected by ultrasound | |
| Two ultrasonic soft markers n = 1078 | CNV anomaly: pCNV n = 63 of 1078 (5.8%)/VOUS n =14 of 1078 (1.2%) | SNP-array n = 77 | ||||||||||
| Three or more ultrasonic soft markers n = 397 | CNV anomaly: pCNV n = 45 of 397 (11.3%)/VOUS n = 4 of 397 (1%) | SNP-array n = 49 | ||||||||||
| Hui Lin et al. [31] | 2020 | Asia | Cross-sectional | 689 | > 34 years old | 16–18 weeks | CMA | Ultrasound soft marker anomalies n = 414 | Abnormal CMA n = 22 of 414 (5.3%) | CMA n = 22 | With patient education and advanced knowledge of phenotype consequences of identified vari ants, prenatal CMA is expected to become an important diagnostic tool in obstetrical practice | |
| Bardin et al. [32] | 2017 | Asia | Retros- pective cohort | 248 | 259 | 32 ±4.4 | Gestational age at diagnosis (weeks) 28.53 ±3.85/gestational age at amniocentesis (weeks) 32.17 ±2.72 | CMA | Late onset abnormal sonographic findings n = 103 | Abnormal CMA/CNV anomaly n = 5 of 103 (4.8%) | CMA n = 95 | if late amniocentesis is undertaken, than CMA should be considered as part of the genetic analysis in cases with abnormal sonographic findings at late second and third trimester |
| Ainsworth et al. [38] | 2017 | America | Retros- pective cohort | 139 | 30.2 (8.2) | NIPS | Major structural abnormalities by organ system n = 77/soft markers n = 62 | Aneuploidy n = 7: TRISOMY 18 AND 21 n = 3 of 7/single gene n = 1 of 9 – low risk: n = 52: Trisomy 18 n = 1 of 52 | NIPS n = 61 | We found that soft markers on ultrasound were associated with the choice of NIPS over amniocentesis, while women who had multiple abnormal ultrasound markers selected amniocentesis. Patients pursuing NIPS after abnormal ultrasound had more soft markers of aneuploidy. | ||
| Lostchuck et al. [33] | 2018 | Australia | Retros- pective | 2494 | 32.2 years (range 15–50) | all trimesters | CMA | Major chromosome abnormalities detected via ultrasound indication n = 341 | CNV anomaly:pCNV n = 75 of 447 (16.7%) – 22q11.2 deletion n = 20 of 447 (4.4%) – Trisomy 21 n = 125 of 447 (27.9%) – Trisomy 18 n = 81 of 447 (18.1%) – Trisomy 13 n = 48 of 447 (1.7%) – Turner (XO) n = 46 of 447 (10.2%) – Other sex chromosome abnormality n = 16 of 447 (3.5%) – Polyploidy n = 21 of 447 (4.6%) | CMA n = 447 | A pathogenic CNV is now the most likely diagnosis after ultrasound-indicated testing, rather than Trisomy 21 or other whole chromosome aneuploidy. Despite steady improvements in first-trimester screening for aneuploidy, the diagnostic yield of ultrasound-indicated tests > 20 weeks has remained stable due to increased utilization of CMAs. Procedures performed for structural abnormalities < 16w continue to have the highest diagnostic yield, supporting the benefits of early fetal structural assessment at 11–13 weeks |
Most studies looked at pregnant women who were referred for sonographic examinations for a variety of reasons; in addition, CMA and SNP arrays gave a higher resolution than karyotyping and are capable of detecting microduplication and microdeletion, which were compatible with the conditions at the time. The cases examined in the included studies were in different trimesters, including the first to third trimesters. We aim to assess each outcome based on the trimester.
In the study by Cai et al. [37] in 2021, prenatal tests like conventional karyotyping and SNP array were performed on 1132 foetuses that confirmed ultrasonic soft marker abnormalities. Depending on the gestational age, different procedures like amniocentesis, chorionic sampling, or cord blood sampling were employed to acquire foetal samples. The mean maternal age was 28.9 years, and the mean number of gestational weeks was 24.3 [37].
In another study conducted by Cai et al. [36] in 2020, foetuses with normal karyotypes were excluded from the study, and so 713 foetuses with abnormalities were selected. The selected foetuses were subjected to CMA, a type of SNP array [36]; subsequently, through this study, the numerous forms of CNVs found by SNP array are categorised into the following groups: benign, VOUS, and pCNVs [36].
A study conducted by Xiang et al. [34] included 5000 samples of cord blood, amniotic fluid, chorionic villi, and foetuses with structural anomalies and soft markers on ultrasonography. It was found that 4022 pregnant women chose both karyotyping SNP and array as screening tools, whereas 978 pregnant women preferred SNP arrays. The mean age of included pregnant women was 32.04 years, and the mean gestational age was 21.28 weeks [34].
Bornstein et al. [30] conducted a retrospective analysis in USA on amniocentesis and chorionic villi sample data acquired in a particular institution between 2010 and 2014. Chromosomal microarray analysis was provided to 3314 patients having invasive genetic testing for various purposes in addition to conventional karyotype. The pCNV prevalence was evaluated between individuals with low-risk indications as well as those with high-risk indications [30].
Beulen et al. [39] conducted research that included all pregnant women with aberrant US results and compared the efficacy of NIPT to invasive diagnostic methods such as CMA and quantitative fluorescence- polymerase chain reaction (QF-PCR). The study did not examine sex chromosomal abnormalities, gender, or foetal fraction. The majority of genetic anomalies discovered were regular whole-chromosome aneuploidies including Trisomy 21, Trisomy 18, and Trisomy 13. Four more aberrant NIPT findings were found, one of which was very suspicious for limited placental mosaicism and one of maternal origin. If the NIPT findings were normal, ultrasound follow-up or newborn checks revealed diagnostic genetic testing within pregnancies (14.7% of 224). Seven instances (3.1% of 224) had clinically significant genetic abnormalities, 2 of which consisted of whole-chromosome aneuploidies including Trisomy 13 and monosomy X [39].
A recent study by Cai et al. [35] in Southern China assessed the utilisation of SNP array evaluation in prenatal care for foetuses with ultrasonography soft indicators and other risk factors including NIPT-positive, abnormal ultrasound structure, chromosomal abnormalities in couples, second trimester serum screening (STSS) high-risk, advanced age, ultrasonic soft marker, and adverse pregnancy history. The study was conducted on data from 8386 pregnancies, dividing them into 7 groups based on various risk factors such as STSS high-risk, ultrasonic soft marker, NIPT-positive, advanced age, adverse pregnancy history, and chromosomal abnormalities in couples. The study included pregnant women aged 18–48 years, with gestational ages ranging 11–36 weeks. The majority of the samples taken (83.1%) were amniotic fluid samples, followed by villi samples (0.7%) and cord blood samples (16.1%) [35].
Bardin et al. [32] investigated the prevalence of chromosomal cytogenetic anomalies among foetuses with late-onset aberrant sonographic findings, discovered that all 103 foetuses had a normal karyotype in an assessment involving women who underwent amniocentesis at or after 23 weeks of gestation for foetal karyotype and CMA investigation, which was recommended due to late-onset abnormal sonographic results. In addition, 95 of these women underwent CMA [32].
In another study, Ainsworth et al. [38] considered patients who received genetic testing following abnormal ultrasonography. Non-invasive prenatal screening or amniocentesis was used for genetic testing. The researchers gathered information on how ultrasound findings were classified, the type of genetic testing done, and the results received. Following an abnormal screening ultrasound, 139 patients were involved in the study and underwent genetic testing. Among these participants, 61 (44%) chose NIPS, whereas 78 (56%) went straight to amniocentesis [38].
Lostchuck et al. [33] investigated the utilisation of ultrasound-indicated chorionic villus sampling and amniocenteses for prenatal diagnostic testing in the Australian state of Victoria. Foetal structural abnormalities, foetal mortality, foetal growth limitation, aberrant genetic – soft markers, amniotic fluid volume, and nonspecific ultrasonography abnormalities were all reasons for these treatments. This study gathered information on the mother’s age, the reason for testing, the type of diagnostic technique utilised, the gestational age, the type of chromosome analysis employed (CMA or G banding karyotype), and the test results [33].
Lin et al. [31] performed a prenatal SNP array analysis on a cohort of prenatal samples in this cross-sectional study. The study aimed to discover chromosomal abnormalities, and the result included 10,377 representatives in total. Chromosomal microarray analysis genetically assessed 689 foetuses in the study; their gestational ages ranged from 16 to 18 weeks, and their mothers were above 34 years old. Chromosomal microarray analysis investigations were ordered to confirm abnormal karyotypes, detect anomalies in ultrasounds, determine advanced maternal age, and relieve parental anxiety. The primary focus was examining homozygosity regions and CNVs, demonstrated by the SNP array [31].
After performing prenatal or postnatal diagnostic tests, QF-PCR was done, and if it was normal, CMA was carried out. Four studies [34, 35, 37, 39] divided women with soft marker sonographic anomalies into groups with single soft marker anomalies or 2 or multiple soft marker anomalies; 2 [30, 31] studies expressed cases into a group with ultrasonic soft marker anomalies generally; one study [38] only discussed about major soft marker anomalies; and one study [32] explained late-onset soft markers.
Soft markers and abnormalities
In karyotyping, anomalies like Trisomy 21, Trisomy 18, and chromosomal structural abnormalities were found in all 3 groups of soft markers (single soft marker, two soft markers, multiple soft markers); rate of Trisomy 21, Trisomy 18, and chromosomal structural abnormalities were, respectively, 12 of 729 (1.6% ), 3 of 729 (0.4%), and 5 of 729 (0.6%) in foetuses with a single soft marker; 7 of 322 (2.1%), 1 of 322 (0.3%), and 3 of 322 (0.9%) in foetuses with 2 soft markers; and 4 of 80 (5%), 1 of 80 (1.25%), and 1 of 80 (1.25%) in foetuses with 3 or more soft markers. Trisomy 13, 47, XXY, 45, X, and mosaic chromosomal number abnormalities were other abnormalities in cases with sonographic soft markers in the study by Cai et al. performed in 2021 [37]. Trisomy 21 and chromosomal structural abnormalities were the most common in foetuses with ultrasonic soft markers, followed by Trisomy 18, Trisomy 13, and other chromosomal number abnormalities [37]. Out of 729 foetuses with a single US soft marker, 322 foetuses with 2 ultrasonic soft markers and 80 with 3 or more ultrasonic soft markers, 26 foetuses, 13 foetuses, and 7 foetuses had a chromosomal abnormality, respectively [37] (Table 1). In the SNP an array of 729 foetuses with a single US soft marker, 322 foetuses with 2 ultrasonic soft markers, and 80 foetuses with 3 or more ultrasonic soft markers; 45 foetuses, 20 foetuses, and 4 foetuses had abnormal CNVs (Table 1). The results of the study by Cai et al. [37] showed no significant difference in the rate of chromosomal abnormalities and abnormal CNVs by karyotyping and SNP array among the ultrasonic soft marker groups [37].
Among 336 foetuses in a study conducted by Cai et al. [36] in 2020 with abnormalities in sonography soft markers, 16 foetuses had abnormal CNVs (16 of 336, 4.7%), 5 (1.4%) of them were pCNVs, and 11 of them (3.2%) were VOUS. Based on the study, abnormal CNVs in foetuses with structural abnormality in sonography were higher than the groups with non-structural abnormality in sonography, which is included ultrasonic soft markers, foetal growth retraction, and amniotic fluid volume abnormality and pericardial effusion [36].
Single nucleotide polymorphism array results of the Xiang study [34] showed 24 of 458 (5.2%) aneuploidy and triploidy, 1 of 458 (0.2%) LOH, and 30 of 458 (6.5%) CNV in groups of foetuses with single soft marker abnormalities, while the results in group of foetuses with multiple soft marker chromosomal abnormalities were found significantly more than the group with single soft markers, including 10 of 72 (13.8%) aneuploidy, and 5 of 72 CNV (6.9%) [34]. The chromosomal abnormality rate in SNP array was lowest in the group with a single ultrasonic soft marker compared to the group with structural anomalies in multiple systems, which was the highest [34].
In the chromosomal microarray analysis of Bornstein et al. [30] involving 1940 foetuses that underwent CMA, it was found that 67% (359/536) of the cases with abnormal sonographic results in the first and second trimesters had soft sonographic markers. Of all the cases with soft markers that underwent CMA, only six had pathological results. Among these pathological cases, 1.1% (4/359) had abnormal CNV, and 0.5% (2/359) were classified as VOUS likely to be abnormal. It was discovered that foetuses with mild abnormalities, such as soft markers, had a decreased prevalence of CMA abnormality compared to those who had significant abnormalities or abnormal NT [30].
Abnormal sonographic results of 73 cases in the study of Beulen et al. [39] among 1940 foetuses that were subjected to NIPT and genetic diagnostic tests were divided into 3 groups: those with one sonographic soft marker, which included 45 out of 73 (61.6%) cases; those with 2 sonographic soft markers, which included 22 cases out of 73 (30.1%); and those with more than 2 soft sonographic markers, which included 6 out of 73 (8.2%) cases. The abnormality diagnosed only consisted of Trisomy 21 from the group with a single soft marker, one case of Trisomy 21 out of 45 (2.2%), and also from the group with 2 soft markers – one case of Trisomy 21 out of 22 cases (4.5%). The abnormality and Trisomy were not detected in the group with more than 2 soft markers, and all 6 cases were normal. The abnormality and Trisomy were not detected in the group with more than 2 soft markers, and all 6 cases were normal. The results of genetic diagnostic tests were similar and confirmed the results of NIPT, including one case Trisomy 21 in the single marker group, and also one case in the group with 2 soft markers, and 2 cases of the total cases (2.6% of the total cases were conducted to confirm NIPT) of Trisomy 21 were among the cases diagnosed with sonographic soft markers [39].
Another study performed by Cai et al. [35] in 2023 showed that the presence of multiple ultrasonic soft markers indicates a greater possibility to have pCNVs [6], which is shown in Table 1: 45 foetuses with pCNVs in the group with 3 or more ultrasonic soft markers (45 of 397, 11.3%), 63 foetuses with pCNVs in the group with 2 ultrasonic soft markers (63 of 1078,5.8%), and 89 foetuses with pCNVs in the group with a single ultrasonic soft marker (89 of 1949, 4.6%) [35].
A study by Bardin et al. [32] involving 103 foetuses with abnormal sonographic findings and normal karyotypes found that the detection rate of abnormal CMA was similar to that of women who underwent amniocentesis due to abnormal early ultrasound findings. However, the detection rate was significantly higher compared to women who underwent amniocentesis and CMA without a medical indication. Out of the 95 women who had CMA performed, 5.3% (5 of 95) had abnormal results, including 5 cases of CNV anomaly, although 5 of 103 (4.8%) had CNV anomaly within abnormal sonographic findings [32].
Among the 139 foetuses identified by ultrasound as abnormal within the investigations of Ainsworth et al. [38], 61 cases were subsequently examined by NIPS; 9 out of 61 (14.7%) tests were pathological; one out of a total of 9 involved a single gene defect; 7 out of 9 cases out of 61 (11.4%) were aneuploid; and out of 7 aneuploid cases, 3 (4.9%) had Trisomy 18 and 21. At the same time, 52 people were included in the group that underwent NIPS examination. Only one (or 1.16%) of the 61 individuals in the low-risk group had Trisomy 21 (Down syndrome). It was concluded that a total of 4 individuals (6.5%) among those assessed with NIPS had Trisomy 18 and 21 and, overall, 40 (65.5%) of the total cases analysed with NIPS had soft sonographic markers [38].
In the study by Lostchuck et al. [33] CMA genetically examined 447 foetuses out of a total of 2494 foetuses; of these, 341 (76.2%) had sonographic markers and abnormalities, 75 (16.7%) had pCNV, 20 (4.4%) had a 22q11.2 deletion, and 125 (27.9%) had Trisomy 21 (Down syndrome). Trisomy 18 represented 81 out of 447 instances (18.1%), Trisomy 13 represented 48 out of 447 (1.7%), Turner (XO) represented 46 cases (10.2%), and polyploidy represented 21 cases (4.6%) [33].
A total of 689 foetuses were used in the study conducted by Lin et al. [31], and 414 of them (or 60%) showed sonographic markers and abnormalities. Additionally, 22 of the 414 foetuses (or 5.3%) had pathological CMA test results [31].
As shown in the studies, the presence of abnormal ultrasonic soft markers alone cannot indicate the existence of chromosomal abnormalities, and more tests are needed to confirm the existence of abnormalities. Ultrasonic soft markers can be considered normal, or by pregnancy progress, they may disappear, but they can be related to chromosomal abnormalities and pCNVs also [6]. For genetic abnormality diagnosis, the CMA method is recommended in foetuses with abnormal sonography, especially in the second or third trimester [36]. The presence of thickened nuchal translucency, short femur, absent nasal bone, ventriculomegaly, and choroid plexus as ultrasonic soft markers should be considered in more and more tests like karyotyping. Single nucleotide polymorphism array must be done because, according to Cai’s study [37], the abnormalities are not limited to chromosomal abnormalities, and they can be abnormal CNVs, so besides normal karyotype, SNP array is recommended so that other abnormalities are not missed.
Trisomy and copy-number variations
Among the genetic abnormalities detected by complementary genetic tests after ultrasound screening and considering the number and type of sonographic soft markers, the most common abnormalities are aneuploidy and triploidy, and the most common abnormality detected among Trisomies is Trisomy 21 [33, 37, 38], followed by Trisomy 18, Trisomy 13, Turner (XO), 47, XXY, mosaic chromosomal number abnormalities, and chromosomal structural abnormalities, and even in one study, abnormalities such as 22q11.2 deletion was seen [33, 37, 38]. Among the studies that have examined both CNV and chromosomal abnormalities, trisomies have been significantly more common than pCNV and VOUS according to studies in which genetic tests have detected both types of abnormalities, including CNVs and Trisomies. Trisomies were detected more often among the studies through SNP array method than the studies that were detected by CMA.
Among studies that examined only abnormal types of CNVs, genetic tests detected the most pCNVs, followed by VOUSs. Pathological cases of CNV and its anomalies are mostly detected by CMA method [30–33, 35–37].
Discussion
Different ultrasonic soft markers are able to be identified during ultrasound examination considering the rapid development of high-resolution ultrasonography as well as prenatal ultrasound diagnostic procedures [25]. Thickened nuchal translucency, plexus cyst, absent nasal bone, hyperechogenic bowel, pyelectasis, choroid ventriculomegaly, short femur, mild tricuspid regurgitation, echogenic intracardiac focus, single umbilical artery, and other ultrasonic soft markers are examples [40–44]. Using ultrasonic soft markers, the studies discovered higher risk of chromosomal abnormalities among foetuses. Nevertheless, ultrasonic soft markers mostly relate to some non-specific index that does not fully reveal the structural abnormalities of the foetus, which might indicate a normal variation [45]. Trisomies 21, 18, and 13, Turner syndrome, and triploidy are the 5 main chromosomal aneuploidies that are associated with the ultrasonography soft markers [46, 47]. Use of the soft markers may increase the positive predictive value in patients with first-trimester combined screening (combination of maternal age, biochemical screening tests of free β-hcg and PAPP-A, and nuchal translucency) [48].
Chromosomal microarray analysis enables a high-resolution evaluation far exceeding that of traditional cytogenetic analysis, and it can detect CNVs associated with known genetic syndromes or abnormal clinical phenotypes. Chromosomal microarray analysis has been found to have an additive value in foetuses with a normal karyotype, in other words prenatal CMA detected clinically relevant CNVs in foetuses with a normal karyotype because it can detect CNVs that may be associated with adverse pregnancy outcomes [30]. Foetuses with ultrasound-detected abnormalities in late pregnancy and normal karyotypes were analysed, and 8.0% (57/713) of the cases had abnormal CNVs. The detection rate of abnormal CNVs in foetuses with sonographic structural malformations (12.7%, 30/237) was significantly higher (p = 0.001) than that in the foetuses with non-structural abnormalities (5.7%, 27/476) [36].
Across several soft marker groupings, Trisomies 21, 18, and chromosomal structural abnormalities have been commonly seen. Trisomy rates varied, ranging from 1.6 to 0.4%, and 0.6% regarding a single soft marker to 2.1%, 0.3%, and 0.9% within 2 soft markers to 5%, 1.25, and 1.25 within at least 3 markers. There were additionally several chromosomal abnormalities such as Trisomy 13, mosaic abnormalities, and 47 XXY. Findings revealed both CNVs and chromosomal abnormalities among foetuses with soft markers, underscoring the need to integrate ultrasonography with microarray approaches. Interestingly, karyotyping and SNP array analyses of various soft marker groups revealed comparable rates of anomalies. The importance of microarray analysis in circumstances with soft markers has been shown by various studies. The outcomes demonstrated that in contrast to non-structural abnormality groups, foetuses having structural abnormalities through sonography, consisting of soft markers, had an increased prevalence of abnormal CNVs. Microarrays were efficient in identifying aneuploidy, Trisomies, and CNVs. The number of soft markers, as well as the kind of genetic test applied, had an impact according to the detection rate. Additionally, it was claimed that in situations with sonographic soft markers together with abnormal nuchal translucency, CMA may successfully detect abnormalities.
In the study by Xiang et al. [34], SNP array analysis of 5000 samples found that 12.3% had abnormalities and 2.6% had clinically significant CNVs, and SNP array identified clinically significant submicroscopic CNVs in foetuses with anomalies on ultrasonography (4.5%), advanced maternal age (1.6%), abnormal maternal serum screening results (2.5%), abnormal NIPT results (2.9%), and other indications (3.0%). Likewise, a minor but not statistically significant increase in the odds of clinically important CNV was found in foetuses with at least one ultrasound soft marker in an American study by Angras et al. [49] in 2020, which is consistent with the study by Xiang et al. [34], who found no statistically significant distinction between the rates of clinically relevant submicroscopic CNVs among foetuses with abnormalities of just one ultrasonic soft marker and more than one ultrasonic soft marker. Meanwhile, when compared to karyotyping, SNP array could detect all aneuploidy and triploidy but not balanced structural chromosomal abnormalities and low-level mosaicism [34]. Additionally, to analyse every one of the 24 chromosome aneuploidies in 57,204 pregnancies, the clinical practical effectiveness of NIPT was evaluated in 2019 by Xue et al. [50], and the results revealed that NIPT performed well in determining T13, T18, and T21. However, the accuracy rate for detecting uncommon foetal chromosome aneuploidies seemed insufficient [50].
Forty-six foetuses with ultrasonic soft markers had chromosomal abnormalities, which were identified by conventional karyotyping. Single nucleotide polymorphism array analysis identified an additional 6.1% (69/1131) of abnormal CNVs in foetuses with ultrasonic soft markers in a study by Cai et al. performed in 2021 [37]. No significant difference was found in the rate of abnormal CNVs among foetuses with one, 2, or 3 or more ultrasonic soft markers [37]; therefor,e the SNP array can fully complement conventional karyotyping in foetuses with ultrasonic soft markers, improving the detection rate of chromosomal abnormalities, which was demonstrated in similar studies [21, 29, 34, 36, 37, 51–56]. Additionally in a meta-analysis and prospective cohort study conducted by Shuyuan et al. [12] in 2020, it was implicated that the foetuses displaying US soft markers should be administered chromosomal microarrays in the second trimester [12]. However, through a related study [35], which recommended that the SNP array should be performed on pregnant women when multiple ultrasound soft markers are detected by ultrasound [35, 44, 57, 58], SNP array can detect not only CNVs, but also uniparental disomy and chimera [36]. Also, other previous studies confirmed the aforementioned point and mentioned that the SNP array is a quick, cost-effective, and trusted method for screening whole-genome uniparental disomy [59, 60], such as in MDS cases in a study by Heinrichs et al. [61], and it has been proven to be a powerful diagnostic tool in cases with developmental delay. Hence, it is suggested that CMA should be considered for genetic analysis in cases with abnormal ultrasound findings in the second and third trimesters of pregnancy [36], then as well other studies which were confirmed that if late amniocentesis is undertaken, than CMA should be considered as part of the genetic analysis in cases with abnormal sonographic findings at late second and third trimester [24, 32, 62–64]. On the other hand, the method was being compared to the amniocentesis method; 44% of patients underwent NIPS and 56% underwent amniocentesis after abnormal ultrasound. Patients electing for amniocentesis had more cardiac, neurological, and gastrointestinal malformations, while soft markers for aneuploidy prompted more NIPS screening. 85% of the NIPS group had negative results compared to 60% of the amniocentesis group [38]. On the other hand, in a similar study by Mardy et al. conducted in 2016, the point declared that false positives can occur with NIPT [65], and in the second and third trimesters of pregnancy, low-risk pregnant women can undergo the advanced screening test NIPS. within other comparable studies by Yunyun et al. [66] and Zhu et al. [67].
Chromosomal microarray analysis offers a more detailed evaluation than traditional cytogenetic analysis and can detect CNVs associated with genetic syndromes or abnormal clinical phenotypes [30, 36]; additionally, one of the main outcomes of a similar study by Lu et al. during [68] as a powerful clinical diagnostic tool, CMA enables fast and accurate diagnosis of mosaic abnormalities and genomic imbalances as the reason for birth defects among neonates [68]. In an individual study analysing foetuses with late ultrasound-detected abnormalities and normal karyotypes, 8.0% of cases were found to have abnormal CNVs [36], which is comparable to a previous study performed by Lu et al. [69] in 2007, regarding 8.5% of patients, CMA found clinically significant genetic abnormalities. The detection rate of abnormal CNVs was significantly higher in foetuses with sonographic structural malformations than in those with non-structural abnormalities [36], then as well studies by Deng et al. [70] and Cai et al. [36].
The studies [1, 6] showed that the rate of abnormal CNVs did not significantly differ among foetuses with different numbers of ultrasonic soft markers [35, 37]. Single nucleotide polymorphism array can detect not only CNVs but also uniparental disomy and illusion, making it a powerful diagnostic tool for cases with developmental delays [36]; in association with various studies [71–74] that demonstrated that the technique of array CGH can be employed to identify copy number variants correlated with intellectual impairment and developmental delay. Likewise, CMA should be considered for genetic analysis in cases with abnormal ultrasound findings in the second and third trimesters of pregnancy [36]. A study comparing NIPS and amniocentesis found that patients opting for amniocentesis had more cardiac, neurological, and gastrointestinal malformations, while soft markers for aneuploidy prompted more NIPS screening [38]. Furthermore, through a similar study by Larion et al. [75], it was presented that the high-risk patient population accepted NIPT quickly, and the positive predictive value for NIPS for aneuploidies except Trisomy 21 was lower due to their higher false-positive rates and also the higher false-negative rate in the study by Meck et al. [76] in 2015. It was suggested that NIPT should not be recommended for the genetic evaluation of the aetiology of ultrasound anomalies because both resolution and sensitivity or negative predictive value are inferior to those of conventional karyotyping and microarray analysis [39]. In contrast, soft ultrasound markers were associated with the choice of NIPS over amniocentesis, while women with multiple abnormal ultrasound markers selected amniocentesis [38]. Patients pursuing NIPS after abnormal ultrasound had more soft markers of aneuploidy [38]. Also, the study by Beulen et al. [39] suggested that NIPT should not be recommended for genetic evaluation of ultrasound anomalies due to lower resolution, sensitivity, and negative predictive value compared to conventional karyotyping and microarray analysis [39]. Another study from 2016 by Lo et al. [77] implied that the most significant chromosomal rearrangements are only possible with conventional NIPT. The most common copy number variants found in the study were deletions and duplications of chromosome regions [31], similarly to the studies by Tucker et al. [78] and Mefford et al. [79]. A pathogenic CNV is the most likely diagnosis after ultrasound-indicated testing, rather than Trisomy 21 or another whole chromosome aneuploidy. Despite steady improvements in first-trimester screening for aneuploidy, the diagnostic yield of ultrasound-indicated tests at > 20 weeks has remained stable due to increased utilisation of CMAs. Procedures performed for structural abnormalities < 16 weeks continue to have the highest diagnostic yield, supporting the benefits of early foetal structural assessment at 11–13 weeks [33]. Soft markers on ultrasound were associated with the choice of NIPS over amniocentesis, while women with multiple abnormal ultrasound markers opted for amniocentesis [31]. Additionally, patients who pursued NIPS after abnormal ultrasound had more soft markers of aneuploidy [31]. Single nucleotide polymorphism array analysis, in combination with karyotyping, can identify clinically significant submicroscopic CNVs in prenatal diagnosis [30, 34], furthermore, CMA enables a high-resolution evaluation surpassing traditional cytogenetic analysis, and can detect CNVs associated with known genetic syndromes or abnormal clinical phenotypes [30, 34]. Also, CMA has been found to have an additive value in foetuses with a normal karyotype because it can detect CNVs that may be associated with adverse pregnancy outcomes [30, 34], as was seen in the outcomes of previous studies by Su et al. [54], Hillman et al. [24], Akalin et al. [80], and Lund et al. [81].
With patient education and advanced knowledge of the phenotype consequences of identified variants, prenatal CMA is expected to become an essential diagnostic tool in obstetric practice [31]. Single nucleotide polymorphism array could additionally identify clinically significant submicroscopic CNVs, and we recommend the combination of SNP array analysis and karyotyping in prenatal diagnosis [34].
Conclusion
Clinical implementation needs to be thoughtfully planned in order for microarrays and ultrasound soft markers to be appropriately included in standard prenatal care. The proper conditions for their utilisation should be defined with obvious guidelines and approaches for mixing various modalities. To expedite processes and provide the best patient outcomes, cooperation between sonographers, obstetricians, geneticists, and a variety of related experts is essential.
In conclusion, ultrasonic soft markers can be normal or related to chromosomal abnormalities which accordance with studies, Trisomy 21, Trisomy 18 and chromosomal structural abnormalities were the most common chromosomal abnormalities related to soft markers. Chromosomal microarray analysis due to its high resolution and ability to detect microdeletions and microduplications is recommended as well as karyotyping, especially in foetuses with normal karyotypes. The ability of SNP array to detect not only CNVs, but also uniparental disomy and chimera, and being a powerful diagnostic tool in cases with developmental delays, makes it worth using. Therefore, based on the results of the present study, it is recommended that genetic tests such as karyotype, SNP array, and CMA be used in foetuses with ultrasonic soft markers, especially in cases with multiple soft markers (2 or more) to detect foetal chromosomal abnormalities. With further progress and investigation on ultrasound soft markers and their relationship with abnormality in the field of pregnancy and screening, it will be possible to use diagnostic and expensive tests only for cases with risk and high probability of abnormality, and in this way, it is possible to avoid the cost and the psychological burden to families and the health care system in future.