Eosinophilic oesophagitis (EoE) is a chronic T2 inflammatory disease with significant eosinophil infiltration restricted to the oesophagus. The understanding of EoE’s underlying mechanism has significantly advanced since the development of differential diagnosis from gastroesophageal reflux disease (GERD) [1]. In general, the EoE pathomechanism is characterized as the interplay of 1) environmental factors assuming premature delivery, antibiotic exposure during infancy, 2) food allergy [2], 3) genetic predispositions with male sex predominance (a 3 : 1 male : female ratio, strong familial associations, and gene abnormalities: CCL26, TSLP, CAPN4, FLG, DSG1) [3], and 4) immune activation partially related to disruption of epithelial barrier integrity and followed by tissue remodelling [4]. More specifically, food antigens and/or cross-sensitized aeroallergens with proteolytic activity may trigger thymic stromal lymphopoietin (TSLP), interleukin (IL)-33, and IL-25 expression in the oesophageal epithelium, leading to dendritic cell (DC)-mediated Th2 cell polarization and their subsequent accumulation in the oesophagus [5, 6]. Notably, preclinical mouse models indicate the indispensable role of IL-33 in type 2 cytokine (IL-5, IL-13) induction and enormous eosinophilic infiltration in the oesophagus [3]. Therefore, the biopsy results confirming increased eosinophil counts in the intraepithelial oesophagus area still remain the gold standard for EoE diagnosis [7].
According to International Consensus Guidelines (2018), the current EoE diagnosis requires identification of oesophageal dysfunction symptoms and detection of eosinophil infiltration in the oesophagus, with at least 15 eosinophils per high-power field [8]. This is followed by applying criteria to exclude systemic and local EoE-related symptoms that might result from other disorders, including gastroesophageal reflux disease (GERD), eosinophilic gastroenteritis, and Crohn’s disease [9]. Thus, achieving at least 90% sensitivity in conventional EoE histology-based diagnosis requires 5-6 biopsy samples from both the distal and proximal oesophagus [1]. Therefore, diagnostic tools enabling precise and differential diagnosis of EoE in clinical routine are required [10, 11].
The development of the EoE Diagnostic Panel (EDP) advanced by Blanchard et al. and Wen et al. and further studies represent the first molecular diagnostic tool targeting 94 genes allowed for the differentiation of active and non-active EoE in paediatric and adult patients based on oesophagus biopsy samples [11, 12]. More specifically, 77 genes were significantly dysregulated in individuals with EoE, with 50 genes being upregulated and 27 downregulated. The molecular panel of genes was specified into 5 categories, namely 1) cell adhesion (CDH16, DSG1, CLDN10, CTNNAL1, CHL1), 2) epithelial-related (FLG, UPK1A, SPINK7, CRISP3, MUC4), 3) inflammation (TNFAIP6, ALOX15, ARG1, MMP12, IGJ), 4) remodelling (POSTN, KRT23, COL8A2, CTSC, ACTG2), 5) eosinophil/mast cell (CLC, CCR3, TPSB2/AB1, CPA3, CMA1), 6) chemokine/cytokine (CCL26, CXCL1, IL4, IL5, IL13). Notably, the gene expression of CCL26, TNFAIP6, and ALOX15 was mostly upregulated, whereas CRISP3, DSG1, and CRYM were found to be downregulated. Importantly, the EDP scoring system correlates with eosinophil counts in epithelial areas and differentiates EoE from GERD. Moreover, the correlation of EDP scores with histological and endoscopic results allows for the distinction of three EoE variants: EoE-like oesophagitis, lymphocytic oesophagitis, and non-specific oesophagitis. These variants are characterized by different compositions of immune cell infiltrates and a lack of eosinophil infiltration. To this end, the development of novel diagnostic tools is required to distinguish conventional EoE in active form, histological remission state, and control individuals, while differentiating the mentioned EoE variants and enhancing diagnostic precision. In this context, Gueguen et al. introduced a novel transcriptomic panel for EoE named the Histologically Active EoE Diagnostic Panel (HAEDP) [13].
The study utilized analysis of bulk RNA sequencing acquired from oesophagus biopsies and clinical data collected from patients included in the Swiss Eosinophilic Oesophagitis Cohort Study (SEECS). Based on the pathologist evaluation, individuals were divided into the following categories: histological inflammatory remission with 1) endoscopically/histologically diagnosed fibrosis (EoE RF+), and 2) without fibrosis (EoE RF–); histologically active EoE 3) with fibrosis (EoE AF+), and 4) without fibrosis (EoE AF–); and 5) healthy controls (Fig. 1). Firstly, the authors found significant differences in transcriptomic profiles in patients with histologically active EoE compared to histological remission and controls. Notably, gene expression patterns did not differ among individuals with or without fibrosis regardless of histological inflammatory status. Samples collected from EoE AF+ patients showed 411 differentially regulated genes (DEGs) compared to controls. Importantly, comparative analysis between EoE RF– and EoE AF– patients, as well as EoE RF+ and EoE AF+ patients, revealed a total of 252 DEGs in the EoE RF– and EoE AF– group, and 246 DEGs in the EoE RF+ and EoE AF+ group. Moreover, patients with the active form of EoE were clearly separated into two clusters, named EoE1 and EoE2. Surprisingly, despite substantial upregulation in expression of the gene encoding eosinophil chemoattractant CCL26 in subgroup EoE2 compared to EoE1, this clustering was not directly linked to eosinophil counts. This analysis also highlighted 46 upregulated and 7 downregulated genes common to all comparisons. Overall, the authors identified 53 dysregulated genes linked to EoE inflammation, providing new insights into the disease’s molecular mechanisms. However, the correlation of clustering with disease activity score may indicate the potential clinical relevance of subgrouping. Moreover, the HAEDP allows us to effectively distinguish patients with an active form of the disease from remission regardless of the fibrosis status and the biopsy site. Given that conventional EoE diagnosis involves invasive procedures, it is important to develop less invasive alternatives with comparable specificity. Potential approaches might be based on measurements in the plasma of blood-based biomarkers for EoE, namely CLC/GAL-10, ECP, EDN, eotaxin-3, and MBP-1 [14, 15].
Fig. 1
Graphical summary of differential diagnosis approaches in eosinophilic oesophagitis (EoE) variants from gastroesophageal reflux disease (GERD) and other EoE-like diseases. Combining a Histologically Active EoE Diagnostic Panel (HAEDP) with an EoE Diagnostic Panel (EDP) as a novel diagnostic tool may make it possible to distinguish patients with an active form of EoE from remission regardless of the fibrosis status. Heatmap represents the genes differentially regulated among investigated groups, namely GERD, EoE RF– (inflammatory remission, no fibrosis), EoE RF+ (inflammatory remission, fibrosis), EoE AF+ (active inflammation, fibrosis), EoE AF– (active inflammation, no fibrosis)
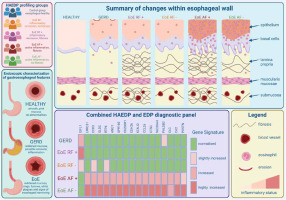
To optimize the diagnostic accuracy of the HAEDP, the newly identified pattern of genes was combined with previously determined EDP. A total of 17 genes were common for both panels with 16 upregulated and only 1 gene with downregulated expression. The expression of genes LRRC31, CCR3, GLDC, RTP4, ANO1, GPR160, C200R1, CDH26, KCNJ2, CCL26, and CTSC was significantly higher in the histologically active EoE, while only IGFL1 remained downregulated in EoE active patients compared to remission (Fig. 1). Those findings may indicate that combining HAEDP and EDP panels may further improve the accuracy of the diagnostic potential in better classifying the patients with EoE. On the other hand, the authors identified 24 genes uniquely upregulated in EDP and 36 in HAEDP, while the expression of 20 and 6 genes, respectively, was downregulated. Unique deregulations in the expression of a total of 36 genes in HAEDP and 60 genes in EDP may indicate the complementarity among the two panels. Thus, the panel advanced by Gueguen et al. might be an accurate tool to help distinguish EoE from other disorders characterized by EoE-related symptoms. However, given the substantial EoE heterogeneity comprising distinct histological inflammatory status, fibrosis development, and epithelial infiltrate composition, the implementation of panels differentiating distinct EoE variants is warranted [7]. The HAEDP holds valuable potential for actively monitoring disease progression. Its clinical significance lies in enabling tailored treatment adjustments by tracking the patient’s current disease state. This capability allows for the development of personalized therapies, better adapted to individual patient needs, reducing the risks of long-term consequences associated with EoE. By utilizing gene expression profiles, this approach can optimize treatment strategies and ensure more effective management, ultimately improving patient outcomes.
To address this issue, the authors utilized recently published data concerning the oesophageal biopsies collected from patients suffering from EoE-like oesophagitis, lymphocytic oesophagitis, non-specific oesophagitis, and conventional EoE, while GERD patients were used as controls. Combining HAEDP and EDP panels differentiates biopsies collected from conventional EoE patients from GERD, EoE-like disease, lymphocytic EoE, and non-specific EoE and controls [10]. The authors advanced a panel of genes, namely CCR3, CTSC, CD200R1, POSTN, CCL26, KCNJ2, GLDC, CDH26, ANO1, and CLC, as a potential diagnostic tool or biomarker set to distinguish EoE variants from other related abnormalities (Fig. 1).
Combining EDP with the recently described HAEDP might be a reliable tool for differential diagnosis of conventional EoE and its variants from EoE symptom-related diseases [10]. However, given the substantial EoE heterogeneity, the number of tested patients using an established panel should be increased [9]. Therefore, a large-cohort, multicentre study is required to estimate the accuracy of the tools. Moreover, better standardization of oesophagus biopsies may be crucial to increase data reliability [16]. Nevertheless, research on improving EoE diagnostic tools and biomarker identification is necessary. We look forward to further developments.


