Introduction
Coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been declared a pandemic by the World Health Organization (WHO). Since its outbreak in December 2019, the disease has affected approximately 235.57 million people worldwide, resulting in approximately 4.81 million deaths globally as of October 3, 2021 (Dong et al., 2020; Sachan et al., 2021; Sokouti et al., 2021). In the fight against this virus, several therapeutic and vaccination approaches have been considered (Akhtar et al., 2020; Baruah and Bose, 2020; Dong et al., 2020; Joshi et al., 2020). According to the WHO, over 123 vaccine candidates are currently in the clinical phase, and 194 vaccines are in preclinical development. A total of 76 different candidate vaccines have been administered; vaccines developed by Pfizer (BNT162b2), SII/Covishield by AstraZeneca/Oxford (AZD1222), Janssen (Ad26.VOC 2.S), Moderna (mRNA 1273), and Sinopharm have been endorsed by the Food and Drug Administration (FDA) (Anderson et al., 2020). As of October 3, 2021, an estimated 6 312 million doses of vaccine have been administered, accounting for 34% of the world’s population (Holder, 2021). Data from the GamCOVID-Vac Phase 3 clinical trials (also known as Sputnik V) were recently revealed, which showed that the vaccine can provide strong immunity to participants infected with SARS-CoV-2 (Logunov et al., 2020). These vaccines are effective in developing immunity from the virus. However, other issues regarding booster doses, immunization regime optimization, correlation with protection, safety, and increased surveillance need to be addressed to further improve vaccine effectiveness (Kim et al., 2021).
Several antiviral therapies are under consideration and require official approval from regulatory authorities (Pinho, 2021). Remdesivir has been authorized for use in the treatment of COVID-19 by the European Medicines Agency (EMA) and the United States Food and Drug Administration (USFDA) (FDA, 2020; Pinho, 2021). Similarly, antibody therapies developed by different pharmaceutical companies are also under review by the EMA for their possible use in the treatment of COVID-19 (Pinho, 2021). Nutraceuticals such as ferulic acid, lipoic acid, monoclonal antibody TJM2 (a neutralizing antibody against the stimulation factor in human granulocyte and macrophage colonies), and plasma therapies have also been investigated as a possible treatment regimen for SARS-CoV-2 (Cunningham et al., 2020; Piyush et al., 2020; ClinicalTrialsArena, 2020; Zhong et al., 2020). It remains vague whether these therapies will be safe, affordable for the masses, and readily available. It is therefore important to continue searching for alternative therapies for COVID-19. Such a strategy may be the re-use of antiviral drugs and small molecules. The reuse of pharmaceutical drugs is the process of finding a new use of an existing drug for entirely different diseases (Medina-Franco et al., 2015; Sultana et al., 2020; Elmezayen et al., 2020). This approach can be an economically viable and rapid method of drug discovery, as the usual process of drug discovery takes several years and costs millions of US dollars (Nishimura and Hara, 2018; Sultana et al., 2020; Elmezayen et al., 2020). Therefore, in the present study, we assessed commonly used drugs and small molecules for their safety and bioactivity. We studied them for treating COVID-19 by focusing on the RNA-dependent RNA polymerase (RdRp) protein using in silico methods. Mangiferin, Flinderole B, Drummondin E, Butanoic acid, 1,7-dihydroxy-3-methoxyxanthone, Friedelin, Benz(a)anthracene, 1,12-dimethyl, Lersivirine, Chloroquine phosphate and Remdesivir were evaluated in this study. Chloroquine phosphate is an approved antimalarial drug, and Remdesivir is an antiviral drug effective against SARS and Middle East Respiratory Syndrome (MERS), viruses that cause other pandemics (Singh et al., 2020). Various studies have demonstrated the antiviral, antidiabetic, antimicrobial, and antisclerotic effects of Mangiferin in vitro and in vivo (Matkowski et al., 2013). Flinderole B has a history of antimalarial and antitrypanosomal activity (Fernandez et al., 2009; Fernandez et al., 2010). The antibacterial activity of Drummondin E has been reported previously (Jayasuriya et al., 1991). Butanoic acid has been instrumental in treating inflammatory bowel disease (Spina et al., 2007). Friedelin obtained from Azima tetracantha exhibited antipyretic, analgesic, and anti-inflammatory activities in mice models (Antonisamy et al., 2011). Lersivirine is an antiviral drug used for treating human immunodeficiency virus (HIV) (Corbau et al., 2010). These compounds that have already demonstrated various pharmacological activities were studied in this study against the RdRp protein of SARS-CoV-2. Studies have shown that RdRp may be among the most specific targets for RNA viruses (Buonaguro et al., 2020). This is because in RNA viruses, this enzyme is necessary for the replication of viral RNA in host cells, and the inhibition of the RdRp protein thwarts the replication of RNA viruses (Zhu et al., 2020; Mishra and Rathore, 2021). Furthermore, as there are no host cell homologs for the RdRp protein, novel therapies that selectively target the RdRp protein will have fewer off-target effects against human host proteins, which will make the therapy relatively safer (Zhu et al., 2020). Remdesivir has shown antiviral activity against different members of the beta-coronavirus family such as SARS-CoV and MERS-CoV (Gordon et al., 2020). Remdesivir shows antiviral activity against these viruses by inhibiting their RdRp protein (Gordon et al., 2020). Consequently, RdRp was identified as a suitable target protein for analysis in this study. Bioactivity properties, molecular properties, solubility, drug-likeness, and various other parameters of the selected compounds were investigated using bioinformatics tools. Compounds that were predicted to be nontoxic, noncarcinogenic, and nonmutagenic and had a better overall drug resemblance score and ADMET properties were analyzed for their ability to bind to the RdRp protein by using molecular docking. Finally, the stability of the interaction between the compounds and RdRp was assessed through molecular dynamics (MD) simulation studies.
Methods
Prediction of the bioactivity, molecular properties, and drug-likeness of the compounds
The SMILES (simplified molecular-input line-entry system) structures of the 10 compounds (Mangiferin, Flinderole B, Drummondin E, Butanoic acid, 1,7-dihydroxy-3-methoxyxanthone, Friedelin, Benz(a)anthracene, 1,12-dimethyl, Lersivirine, Chloroquine phosphate and Remdesivir) were obtained from PubChem in SDF format (Kim et al., 2018). The SMILES structures of these compounds were then used as input in Molinspiration online server using the default parameters to determine their bioactivity and molecular properties (https://www.molinspiration.com/cgi-bin/properties) (Molinspiration). The SMILES structure of the selected compounds was provided to the Molinspiration webserver as input. This server helps to predict various molecular properties such as n-octanol-water partition coefficient (log P), number of atoms (Natom), molecular weight (MW), number of hydrogen bond donors (nON), number of hydrogen bond acceptors (nOHN), and total polar surface area (TPSA). These properties are used to evaluate whether a compound is likely to be an active drug molecule in humans based on Lipinski’s rule of five (Lipinski et al., 2001). Molecules violating Lipinski’s rule of five may have problems with bioavailability. This server uses a sum of fragment-based contributions and correction factors to calculate the logP value. Similarly, the TPSA is calculated by this server based on the summation of tabulated surface contributions of polar fragments. This server also helps to predict various bioactivities of the compounds, such as kinase inhibitor (KI), protease inhibitor (PI), or enzyme inhibitor (EI). It also helps to determine whether the compounds are ionchannel modulators (ICMs), G protein-coupled receptor ligands (GPCRs), or nuclear receptor ligands (NRLs). Finally, the drug-likeness score was predicted by the druglikeness and molecular property prediction online server using default settings available at Molsoft (https://www.molsoft.com/mprop/) (Molsoft). The SMILES structures were provided as input to the Molsoft webserver. This server predicts overall drug-likeness by using Molsoft’s chemical fingerprints that are trained using 5000 marketed drugs and 10 000 nondrug compounds. The server also predicts various molecular properties such as water solubility, molecular polar surface area, and octanol/water partition coefficient using different machine learning methods such as PLS-regression and Random Forest regression.
Prediction of the solubility and toxicity of the compounds
Osiris Property Explorer software with default settings was used to determine the toxicity and solubility of the molecules. This software predicts whether the compound is irritant, hepatotoxic, or mutagenic and whether it affects reproductive health. It predicts these properties based on the functional group similarity of query molecules with in vivo and in vitro validated molecules in its database (Ayati et al., 2012). If the compounds are toxic or nonsoluble, the results will appear in red, and if the compounds are soluble and nontoxic, the results appear in green. The solubility of the drug is predicted based on the values of logS. The compound with a higher logS value has low solubility and vice versa. A compound with higher aqueous solubility will have better absorption and distribution (Bhati et al., 2019). The SMILES structure of the compounds was provided to the software as input, and the software was run at default settings.
Prediction of ADMET properties
For a compound to be successful under clinical trials, it must have good absorption, distribution, metabolism, and excretion (ADME) properties. Moreover, it must be nontoxic. Therefore, for the prediction of the ADMET properties, admetSAR version 2.0 webserver was used (http://lmmd.ecust.edu.cn/admetsar2/) (Yang et al. 2018). The admetSAR2 webserver provides information about human intestinal absorption of compounds, the blood-brain barrier crossing ability of the compounds, AMES toxicity, hepatotoxicity, and carcinogenicity of the compounds. The admetSAR2 server implements various machine learning methods such as random forest, k-nearest neighbors method, and support vector machine methods to build the models to predict the ADMET properties (Yang et al., 2018). The training data for constructing these models are collected from various databases such as ChEMBL, DrugBank, and CPDB along with various published scientific articles (Yang et al., 2018). The admetSAR webserver was used because it includes the most diverse metabolism properties. It also includes the latest and most comprehensive manually curated ADMET information of chemicals. The SMILES structure of the selected compounds was used as input to the web server, and the results were predicted using the default settings.
Molecular docking and MD simulation
To study the interaction between the compounds and the RdRp protein, initially, a molecular docking study was conducted using the PatchDock webserver available at the website: https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php (Schneidman-Duhovny et al., 2005). The PatchDock server is a geometry-based molecular docking algorithm and determines the ligand-receptor interaction based on molecular shape complementarity and atomic contact energy (Schneidman-Duhovny et al., 2005). The PatchDock algorithm first determines the molecular surface of the molecules and detects their geometric patches. Then, geometric hashing and pose-clustering matching techniques are used to determine complementary patches to generate the candidate transformation followed by evaluation of the transformations using a scoring function that considers atomic desolvation energy and geometric fit. Finally, root means square deviation (RMSD) is calculated to remove the redundant solutions (Schneidman-Duhovny et al., 2005). This server was used for molecular docking because it is highly efficient and fast (run time is less than 10 min for the input of two proteins with a mean size of 300 amino acids).
Before docking, the SDF file of the compounds was converted to a PDB file by Marvin-View software (http://www.chemaxon.com/). Then, the PDB file of compounds was used as a ligand and the PDB file of the RdRp protein (PDB ID: 6M71) was used as a receptor. While performing molecular docking analysis using the Patchdock webserver, the complex type protein-small ligand docking was selected, and an RMSD value of 1.5 Å was used. The selected PDB 6M71 for SARS-CoV-2 RdRp was prepped by Discovery Studio 4.1 (DS) automated protein preparation method. The natural product-based small molecules, i.e., Drummondin E and Flinderole B, were pre-processed to create a protonation state by the “Prepare Ligands” method in DS 4.1 at the pH of 7.0 ± 0.5, following which the molecular geometry of the resultant molecules was determined. The Drummondin E-RdRp and Flinderole B-RdRp complexes were further analyzed by MD simulations using a commercially available “Flare” module of Cresset software. Herein, MD simulation was run in the Flare module, which is based on OpenMM package, using AMBER/GAFF2 as force fields and AM1-BCC as the charge method. The solvent system was constructed equivalent to the protein-ligand complex by the explicit solvent water model by keeping the shape of the box as orthorhombic with dimensions of 10 × 10 × 10 Å. The protein-ligand complex was later minimized by setting the minimum energy tolerance at 0.25 kcal/mol and equilibrated for 200 ps before initiating the MD run. The simulations were performed at a time step of 1 fs and 100 ns as recording time.
Results
Prediction of the bioactivity, molecular properties, and the drug-likeness of the compounds
The PubChem ID and the SMILES structures of the selected compounds are provided in Table 1. Out of the 10 selected compounds, 1,7-dihydroxy-3-methoxyxanthone, Butanoic acid, Drummondin E, and Lersivirine did not violate any rule in Lipinski’s rule of five, whereas the other compounds violated 1 or 2 rules (Table 2). The compounds that violated 1 Lipinski’s rule of five were: Benz(a)anthracene, 1,12-dimethyl and Chloroquine phosphate, and the compounds that violated 2 Lipinski’s rule of five were Flinderole B, Mangiferin, and Remdesivir. If a compound follows Lipinski’s rule of five, it has a higher probability of being an active drug-like molecule (Lipinski et al., 2001). The results of the molecular property analysis by the Molinspiration tool are shown in Table 2. The bioactivity of the compounds was also determined by the Molinspiration tool. 1,7-dihydroxy-3-methoxyxanthone was determined to be a NRL, while Benz(a)anthracene was likely to be an EI and KI. Chloroquine phosphate was predicted as a GPCR ligand, ICM, and KI, whereas Drummondin E and Mangiferin were predicted to be an NRL and EI. Flinderole B is likely to be a GPCR ligand and EI, and friedelin is expected to be an NRL and EI. Lersivirine is also likely to be an NRL and KI. Remdesivir showed the most diverse range of bioactivities. It was found to be a GPCR ligand, KI, EI, and PI. The bioactivities of these compounds are listed in Table 3. Finally, Chloroquine phosphate, Drummondin E, Flinderole B, Mangiferin, and Remdesivir were predicted to be potential drug-like molecules based on their drug-likeness score determined by the Molsoft tool. The drug-likeness score of Chloroquine phosphate, Drummondin E, Flinderole B, Mangiferin, and Remdesivir were 1, 0.38, 0.37, 0.25, and 0.13, respectively. The drug-likeness values of the other selected molecules are given in Table 4. Compounds with zero or negative drug-likeness scores were not considered to be drug-like.
Table 1
Compounds considered in the present study
Table 2
Molecular properties based on the Molinspiration online server
Table 3
The bioactivity analysis of tested compounds
Prediction of the solubility, toxicity, and ADMET properties of the compounds
The Osiris Property Explorer tool was used to determine the solubility and toxicity of the selected molecules. Based on the ClogS score, which is used to predict aqueous solubility of compounds, the Osiris Property explorer tool predicted that Benz(a)anthracene (ClogS score = –7.12) and Friedelin (ClogS score = –6.97) had better aqueous solubility than other compounds used in this study because a lower ClogS score implies higher aqueous solubility. All the compounds, except flinderole B and friedlein, were found to be mutagenic, irritant, and tumorigenic and to affect reproductive health. Drummondin E was found to be nonirritant, nonmutagenic, and nontumorigenic, but it could affect reproductive health. The predicted solubilities and toxicities of the compounds are listed in Table 5. For a compound to be successful under clinical trials, it must have good ADME properties. Moreover, it must be nontoxic. Therefore, for predicting the ADMET properties, the admetSAR2 webserver was used (Yang et al., 2018). After the overall analysis of ADMET properties of the compounds, Flinderole B and Drummondin E were predicted to possess the ability to cross the blood-brain barrier. These two compounds were also predicted to permeate through the human intestine and get absorbed there. These two compounds were also found to be noncarcinogenic. The detailed ADMET properties of all the other selected molecules are given in Table 6.
Table 5
Toxicity and solubility of the selected molecules based on Osiris Property Explorer
Table 6
ADMET properties of the analyzed compounds
Molecular docking and in silico studies
The selection of the molecule for further analysis using molecular docking was based on two parameters: its drug-likeness score including ADMET properties (as described above) and its binding efficiency with RdRp. Flinderole B and Drummondin E fulfilled both criteria. Benz(a)anthracene, 1,12-dimethyl, Remdesivir and Friedelin were the best hits based on atomic contact energy (Table 7). Benz(a)anthracene with the lowest atomic contact energy (–268.16 kcal/ml) in comparison to other molecules was the best molecule that could interact with the SARS-CoV-2 RdRp protein out of the 10 molecules used in this study. Lower atomic contact energy implies that the complex will be more stable and favorable due to the low desolvation energy (Guo et al., 2012). However, Benz(a)anthracene was not explored further by MD simulation because of its low drug-likeliness score (–1.33) (Table 4). Similarly, Friedelin was also not considered for further analysis because of its low drug-likeness score (0.43). Remdesivir was used as a positive control in our analysis. As it is already an approved drug, we did not pursue this molecule for further MD simulation analysis.
Table 7
Molecular docking scores of the tested compounds with the SARS-CoV-2 RdRp protein
Flinderole B and Drummondin E exhibited a better drug-likeness score (0.38 and 0.37, respectively) than the other compounds used in this study (Table 4) and the ability to permeate and get absorbed in the human intestine, and they were found to be noncarcinogenic, nonirritant, nontumorigenic, and nonmutagenic. Hence, these two compounds were selected for further analysis as drug targets. These two drugs could bind with RdRp. The PatchDock results showed that the interaction between Flinderole B and RdRp had an atomic contact energy of –116.95 kcal/mol and a geometric shape complementarity score of 6316. Similarly, the interaction between Drummondin E and RdRp had an atomic contact energy –87.69 kcal/mol and a geometric shape complementarity score of 5334. The complexes with the high geometric shape complementarity score have the least steric hindrances and a wide interface area and are more stable (Schneidman-Duhovny et al., 2005). Negative or low atomic contact energy implies the complex is more stable and favorable due to the low desolvation energy (Guo et al., 2012). Overall, in this study, the interactions between Flinderole B and Drummondin E with RdRp showed better atomic contact energy and geometric shape complementarity score than the interactions between other compounds and RdRp. The atomic contact energy and geometric shape complementarity score of the interactions between RdRp and other compounds are shown in Table 7.
The tertiary structure of the SARS-CoV-2 RdRp protein (PDB ID: 6M7I) was prepped by Discovery Studio 4.1 (DS) automated protein preparation method. It was initially pre-processed by adding the missing atoms in incomplete residues. Further pre-processing involved the missing loop modelling, co-crystallized water elimination, and protonation of the titratable residues via the application of the CHARMM force-field. The natural product-based small molecules, i.e., Drummondin E and Flinderole B, were processed to generate the protonation state by the application of DS studio 4.1 “Prepare Ligands” method at the pH of 7.0 ± 0.5, which was followed by calculation of the molecular geometry of the resultant compounds.
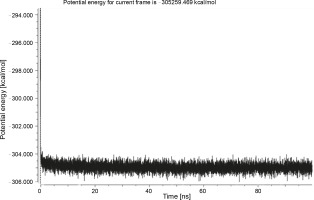
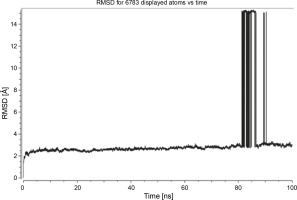
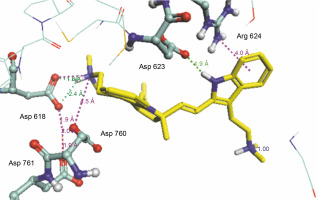
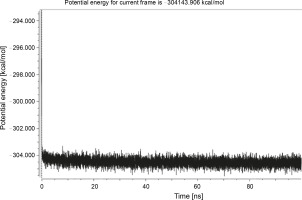
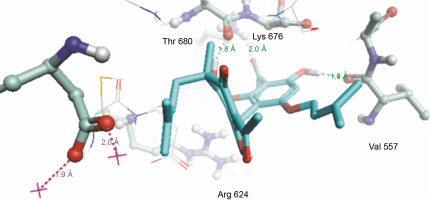
Herein, the active site of the target protein was specified by the “define and edit binding site” tool. The MD simulation was performed for 100 ns with the time step of 1 fs. The stability of the protein-ligand complex was evaluated by considering the RMSD values. RMSD plots corresponding to the Drummondin E and RdRp complex were predicted as fairly stable during the simulation period of 100 ns as shown in Figure 1. The interaction diagrams corresponding to the Drummondin ERdRp complex (Fig. 2) after simulation run time show maintenance of key interactions with Thr 680, Asp 624, Lys 676, and Val 557. These key interactions type along with the percentage of contacts are shown in Table 8. Among these key amino acids, Val 557 (97.1%) and Thr 680 (86.2%) showed the highest percentage of contact retention till the end of the simulations. The same complex was also predicted as stable in terms of potential energy during 100 ns MD simulations (Fig. 3). On the other hand, the RMSD plot corresponding to the Flinderole B-RdRp complex (Fig. 4) revealed that the complex maintained stability for a simulation period of 0–80 ns, while it showed fluctuations within the simulation period of 80–90 ns. This complex was able to regain stability after the simulation period of 90 ns. The interaction diagrams corresponding to the Flinderole B-RdRp complex displayed key interactions with Asp 618, Asp 760, Asp 623, Arg 624, and Asp 761, as presented in Figure 5. The type of interactions between these key amino acids and Flinderole B is explained in detail in Table 9 in terms of a salt bridge, hydrogen bonding, and cation-pi. The N2 atom of Flinderole B formed salt bridges with Asp 618, Asp 623, and Asp 760 residues of the SARS-CoV-2 RdRp protein. Similarly, the H2, H3, and H4 atoms of Flinderole B formed hydrogen bonds with Asp 618, Asp 623, and Ser 681 residues of the SARS-CoV-2 RdRp protein, respectively. The C22 atom of Flinderole B formed cation-pi bonds with Arg 553 and Arg 624 residues, and the C24 atom of Flinderole B formed a cation-pi bond with Arg 624 of the SARS-CoV-2 RdRp protein. The percentage contacts of Flinderole B with different residues represented in Table 9 revealed that Flinderole B showed a 96.6% contact frame with Asp 618. The complex was also predicted to be similarly stable in terms of potential energy as that of the Drummondin E-RdRp complex. The potential energy vs. time frames plot is shown in Fig. 6.
Table 8
Molecular dynamics results corresponding to drummondin E in complex with RdRp in terms of the type of noncovalent interactions and % contacts retained throughout the simulation period of 100 ns
Table 9
Molecular dynamics results corresponding to flinderole B in complex with RdRp in terms of the type of noncovalent interactions and % contacts retained throughout the simulation period of 100 ns
Fig. 1
Graph showing the RMSD value of the Drummondin E-SARS-CoV-2 RdRp complex during the 100 ns molecular dynamics simulation analysis
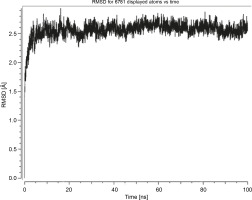
Fig. 2
Simulation interaction diagram of Drummondin in the catalytic sites of the SARS-CoV-2 RdRp protein after the simulation period of 100 ns
Discussion
COVID-19 caused by SARS-CoV-2 has been declared as a pandemic by the WHO. As of October 3, 2021, SARS-CoV-2 has infected 235.57 million people worldwide (WHO 2021). With the advent of a new mutant strain with state-of-the-art proteins, the current FDA-approved vaccines need constant vigilance and may need to reconsidered (Callaway, 2020; Ghaebi et al., 2020; Korber et al., 2020). Currently, the FDA and EMA have approved Remdesivir for emergency use in the treatment of COVID-19, and several other therapies are under investigation and awaiting approval (FDA, 2020; Pinho, 2021).
SARS-CoV-2 is a single-strand positive RNA virus with a genome length of approximately 30 kb (Rastogi et al., 2020). Upon its entry into the host cell, it overtakes the host protein translation system to synthesize pp1a and pp1ab polyproteins (Wu et al., 2020). These polyproteins are cleaved by proteases, namely 3C-like protease (3CL pro) and Papain-like protease (Pl pro), to form functional nonstructural proteins (NSPs): RNA replicase–transcriptase complex (RdRp) or the helicase. The (+) strand is used as a template to produce the genomes of new virions and subgenomic RNAs that are transcribed and translated to produce spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins (Kim et al., 2020; Rastogi et al., 2020). The cryo-electron microscopy-based tertiary structure of the SARS-CoV-2 RdRp protein (PDB Id: 6M71) was released on April 1, 2020 in the public domain (Gao et al., 2020). It is an essential enzyme required for replication and subgenomic RNA amplification. Like other RdRps, it is a conserved protein and has been targeted for developing antiviral therapies against SARS-CoV-2 (Aftab et al., 2020; Gao et al., 2020; Malin et al., 2021). Various drugs that specifically target the RdRp are Remdesivir, Galidesivir, Ribavirin, Favipiravir (T-705), Tenofovir, and Sofosbuvir (Maag et al., 2001; Furuta et al., 2017; Al-Tawfiq et al., 2020; Cao et al., 2020; Elfiky, 2020a; Wang et al., 2020). However, the presence of the exonuclease domain in the nsp14 protein of SARS-CoV-2 could be a challenge in developing nucleoside analog (NA)-based antiviral drugs such as Remdesivir and Galidesivir because the exonuclease domain could excise the incorporated nucleoside, eventually leading to resistance to NA antivirals (Shannon et al. 2020). Hydroxychloroquine (HCQ) has also shown promising results in vitro, and preliminary clinical results seem to be promising; however, the studies are with low confidence (Gautret et al., 2020). Similarly, other drugs such as Lopinavir/ritonavir also did not show promising results in the treatment of patients with severe COVID-19 (Cao et al., 2020).
Therefore, it is important to seek other therapies for COVID-19. One such strategy can be the repurposing of small molecules and antiviral drugs (Andrade et al., 2020; Mohapatra et al., 2020). This approach can be a time saver and can reduce a huge sum of capital investment (Nishimura and Hara, 2018). Therefore, in the present study, we attempted to evaluate the efficacy and safety of drugs and small molecules commonly used against SARS-CoV-2 infection. Remdesivir and Chloroquine phosphate are already approved drugs, and they are used in the present study to test and validate the in silico approach described in the manuscript (Lamb, 2020; Zhang et al., 2020). Among the other molecules, Butanoic acid is already an FDA-approved flavoring agent. Lersivirine is in phase IIb clinical trial against HIV (Platten and Fätkenheuer, 2013). Flinderole B and Drummondin E are already proven drugs with antimalarial and antibacterial activity, respectively (Jayasuriya et al., 1991; Fernandez et al., 2009; Fernandez et al., 2010; Hernández-López et al., 2014). Mangiferin has been suggested for the treatment of infection caused by the porcine reproductive and respiratory syndrome virus and has been patented for its antiviral property in China (Li et al., 2019; Feng et al., 2020). 1,7-dihydroxy-3-methoxyxanthone has been patented in Australia for its antibacterial properties (Moffett and Shah, 2006; Gittins and Trivedi, 2013). Friedelin is hypolipidemic and has antifungal, antibacterial, and anti-inflammatory properties (Mokoka et al., 2013; Duraipandiyan et al., 2016; Nakamura et al., 1997). Friedelin has also been patented for its analgesic property (Nakamura et al., 1997). Benz(a)anthracene,1,12-dimethyl is a potential carcinogenic agents; it is used in the present study as a negative control to validate whether it meets the drug-likeness score of (Dong et al., 2018). It is worth to mention that although Benz(a)anthracene, 1,12-dimethyl; Lersivirine; and Friedelin have good atomic energy contact against the SARS-CoV-2 RdRp, they failed in the drug-likeliness score and thus were not considered in the present study (Table 4 and Table 7).
Among the compounds studied, Flinderole B and Drummondin E exhibited a better drug-likeness score (0.38 and 0.37, respectively) than the other compounds tested in this study (Table 4); furthermore, they showed the ability to permeate and get absorbed in the human intestine and were noncarcinogenic, nonirritant, nontumorigenic, and nonmutagenic as compared to other drugs in this study (Table 5 and Table 6). Drummondin E (C28H34O8) is a derivative of filicinic acid, initially isolated from Hypercium drummondii which is commonly called as St. John’s wort. It is an effective antimicrobial agent and shows activity against gram-positive bacteria (Jayasuriya et al., 1991). Flinderole B (C34H44N4) is reported to have antimalarial activity (Fernandez et al., 2009). Based on computational screening, these two drugs were further evaluated to determine their capacity to interact with RdRp. Molecular docking analysis showed that both these compounds can bind to RdRp. Molecular docking is used to show the possible interactions between the compounds in the target protein’s active site. The molecular docking tool, however, considers the proteins as rigid structures, thereby making the molecular docking process a “rough” approach. Hence, molecular docking was succeeded by MD simulation analysis to characterize the conformation, position, and orientation of the ligands in the target protein’s activity. The MD simulation studies showed that the interactions of both compounds with RdRp were stable. These studies demonstrated that Flinderole B and Drummondin E may be potential inhibitors of the RdRp protein in SARS-CoV-2 and may interfere with its reproductive and transcription processes. Previous studies aimed to identify molecules that could target the SARS-CoV-2 RdRp protein using molecular docking and MD simulation studies used the in silico approach to model the tertiary structure of the SARS-CoV-2 RdRp protein (Elfiky, 2020a; Elfiky, 2020b). However, in our study, we used the tertiary structure of the SARS-CoV-2 RdRp protein obtained from the Protein Data Bank, which was constructed with the guidance of a cryo-electron microscopy map. An in silico study reported that Flinderole B can bind and inhibit nonstructural protein 2 (nsP2) proteases of various alphaviruses (Byler et al., 2016). Additional in silico studies have also identified potential compounds with anti-SARS-CoV-2 potential. Beta-sesquiphellandrene, an antiviral compound found in zinger, has been reported as a potential inhibitor of the spike protein of SARS-CoV-2 in an in silico study where molecular docking and MD simulation studies were performed (Joshi et al., 2020). Similarly, taraxerol a phytochemical isolated from Clerodendrum spp., has also shown the ability to inhibit the RdRp of SARS-CoV-2 in silico (Kar et al., 2020). Another in silico study reported that the C60 fullerene can interact with the catalytic binding pocket of the SARS-CoV-2 RdRp protein (Hurmach et al., 2021). The in silico study also provided insights related to the mechanism of C60 fullerene, where it was predicted to block the RNA synthesis pore and prevent the binding of the Nsp8 cofactor (Hurmach et al., 2021). However, our study has not been able to provide such insights about the possible impacts of interactions of Flinderole B and Drummondin E. In another study that aimed to identify drugs that could target the SARS-CoV-2 RdRp protein, the author identified several FDA-approved drugs such as Sofosbuvir, Ribavirin, Galidesivir, Remdesivir, Favipiravir, Cefuroxime, Tenofovir, and Hydroxychloroquine as potential inhibitors of the SARS-CoV-2 RdRp protein (Elfiky 2020b). In another study, the authors targeted FDA-approved drugs and found that Pitavastatin, Ridogrel, and Rosoxacin could bind to the active site of the SARS-CoV-2 RdRp protein (Baby et al., 2020). In comparison to the study where all the selected compounds were FDA-approved drugs, none of the compounds used in our study, except Butanoic acid and Remdesivir, have been approved by FDA. To date, FDA has not approved Flinderole B and Drummondin E for the treatment of any diseases. Hence, the capacity of these compounds as potential antiviral drugs demands further validation by additional in vitro and in vivo studies.
Conclusions
COVID-19 has been declared a pandemic that has resulted in the death of nearly 5.5 million people worldwide. In the present study, small molecules were investigated as potential drugs for targeting the SARS-CoV-2 RdRp protein. RdRp is a nonstructural protein (Nsp12) that belongs to the nucleotidyl transferase class and is conserved in all beta-coronaviruses. Our molecular simulation analysis showed that two compounds, namely Flinderole B and Drummondin E, interact with the catalytic residues of the protein, thus suggesting that these compounds can block the replication of the virus and hamper its life cycle. In addition, compared to the other compounds used in this study, these two compounds had a better drug-likeness score, had the ability to penetrate and be absorbed in the human intestine, and were nonmutagenic. As new strains of SARS-CoV-2 are emerging, the risk of new outbreaks across the world has also increased. In the present study, the two compounds identified could target the SARS-CoV-2 RdRp protein and limit the replication and growth of SARS-CoV-2. However, the capacity of these compounds as potential antiviral drugs against SARS-CoV-2 has been determined only through in silico studies. Hence, in the future, multiple rigorous and extensive in vitro, in vivo, and clinical studies are required to corroborate the ability of the two compounds, Flinderole B and Drummondin E, to inhibit replication and growth of SARS-CoV-2. Future studies can also aim to identify whether Flinderole B and Drummondin E can interact with the catalytic binding pocket of the SARS-CoV-2 RdRp protein by in silico and in vitro investigations.