Introduction
Living organisms including microorganisms are known to synthesize natural pigments (Rao et al., 2017), which are primarily pharmacologically active compounds. These secondary metabolites confer protection to bacteria against extreme temperature, ultraviolet radiation, and desiccation (Wada et al., 2013). Pigment production is also triggered by microorganisms through quorum-sensing mechanisms to inhibit or kill other microorganisms present in their surroundings (Soliev et al., 2011).
Microorganisms are a preferred source of pigments because of ease of extraction, production manipulation, and availability throughout the year. Microbial pigments are also nontoxic and biodegradable as compared to their synthetic counterparts (Rao et al., 2017). Because of their therapeutic nature, as shown by their remarkable antioxidant, cytotoxic, antimicrobial, and anticancer properties, bacterial pigments have a wide range of applications in pharmaceutical industries (Soliev et al., 2011). Carotenoid pigments play a vital role in protecting bacterial and plant chlorophyll from photo-destructive reactions and are therefore known as photo-protective agents (Sandmann, 2014; Kushwaha et al., 2020). Carotenoids act as antioxidants (Sarker and Oba, 2018a, 2018b, 2020a, 2020b) and are used as vitamin supplements, as their intake prevents photo-aging and sunburns (Numan et al., 2018). Certain quinone-derived pigments demonstrate antimicrobial, insecticidal, and anticancer activities (Saha et al., 2008; Numan et al., 2018).
In the human body, oxidative stress is generated due to the accumulation of free radicals, which is the underlying reason for the development of chronic neurological and cardiovascular diseases and cancers (Phaniendra et al., 2015). Antioxidants possess the ability to destroy free radicals by donating electrons, thereby resulting in their neutralization (Sarker and Oba, 2019a, 2020c, 2020d; Sarker et al., 2020a, 2020b). On the other hand, synthetic antioxidants are not preferable for long-term consumption because of their reported side effects (Phaniendra et al., 2015). Therefore, recent studies on antioxidants have focused on screening different natural compounds and evaluating their pharmaceutical activities (Agustina et al., 2021; Foong et al., 2021; Sakihama and Yamasaki, 2021).
In the present study, various microbial pigments were extracted from indigenous microorganisms isolated from a mangrove rhizosphere habitat. These pigments were characterized by UV-Vis spectroscopy and Fourier Transform Infrared (FTIR) analysis. The pigment production was optimized, and the antioxidant potential of the pigments was determined by chemical assays and an in vitro assay using a murine macrophage cell line.
Materials and methods
Isolation and identification of the bacterial strains
Mangrove soil and water samples (Mumbai; 19.352559°, 72.847327°) were used to isolate the pigment-producing bacteria. The samples were cultured on nutrient agar (M001, HiMedia Laboratories, Mumbai) and incubated for 48 h at 37°C. The obtained pigmented colonies were maintained on sterile nutrient agar slants and in glycerol stocks. These cultures were identified according to their morphological characteristics (Suresh et al., 2004; Vaz-Moreira et al., 2012; Tang et al., 2017) and 16S rDNA sequencing.
DNA extraction of the isolated bacterial cultures
DNA extraction of all the cultures was performed by the phenol-chloroform method (Köchl et al., 2004). To extract DNA, 1 ml of cell suspension from the overnight grown bacterial cultures was centrifuged at 5000 rpm for 10 min to obtain cell pellet, to which 500 μl of 10 μM Tris-EDTA buffer (pH 8.0) and 20 μl Proteinase K (20 mg/ml) were added. The mixture was vortexed, and an equal volume of buffered phenol (pH 8.0) was added; this mixture was vortexed and centrifuged at 14 000 rpm for 10 min. The aqueous phases were collected, followed by the addition of 500 μl of chloroform, vortexing, and centrifugation at 14 000 rpm for 10 min. The aqueous phases were collected once more, and 50 μl of 3 M sodium acetate solution and 800 μl of 100% ethanol were added. This solution was refrigerated at ‒20°C for 1 h. The DNA was recovered by centrifugation at 14 000 rpm for 30 min, and the supernatant was discarded. To wash the pellet, 1 ml of 70% chilled ethanol was added, and the tube was centrifuged at 14 000 rpm for 20 min. The pellet was dried and then dissolved in 50 μl of Tris EDTA buffer (Köchl et al., 2004). The purified DNA was then subjected to polymerase chain reaction (PCR) for amplifying the 16S rRNA gene.
PCR amplification of the purified DNA and 16S rDNA sequencing
PCR was used to amplify the 16S rRNA gene. The primers used for PCR were universal primers: 27F forward primer (5 ˊ‒AGAGTTTGATCCTGGCTCAG‒3ˊ) and 1492R reverse primer (5ˊ‒GGTTACCTTGTTACG ACTT‒3ˊ) (Senthilraj et al., 2016). One microliter of purified DNA (10–100 ng) was added to the reaction mixture containing 12.5 μl Master Mix (EconoTaq, LUC300352, Sigma-Aldrich), 1 μl of each of the primers, and 9.5 μl of double-distilled water (the final volume was 25 μl). The master mix consisted of dNTPs, Taq polymerase, MgCl2, and the reaction buffer. The PCR conditions were as follows: an initial denaturation step of 2 min at 94°C, followed by 35 cycles of 94°C for 1 min (denaturation), 55°C for 1.5 min (annealing), 72°C for 1 min (extension), and then a final extension at 72°C for 3 min (ESCO Thermal cycler, Singapore) (Senthilraj et al., 2016). The PCR-amplified products were visualized on ethidium bromide-stained 1% agarose gel under UV light. The amplified products were gel-purified (HiPurA™ PCR product purification kit, HiMedia, India) and sequenced by the Sanger sequencing method using the ABI 3500 Series Genetic Analyzer platform (Thermo Fisher Scientific, USA). The obtained sequences were analyzed using the NCBI BLASTN (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Gupta et al., 2015) and the Ribosomal Database Project 11 (http://rdp.cme.msu.edu/) (Khaneja et al., 2009). The sequence alignment was performed using Clustal W (https://www.ebi.ac.uk/Tools/msa/clustalo/), and the phylogenetic tree was assembled in MEGA X (https://www.megasoftware.net/).
Optimization of growth conditions for the bacterial isolates
The effect of temperature and salt concentration on bacterial growth was studied by growing the isolates in suspension tubes/96-well plates containing sterile nutrient broth (NB) (MM244, HiMedia Laboratories). The initial culture density of the inoculum was 0.1 OD (5% v/v). The optimum growth temperature of these isolates was determined by incubating the cultures at 10, 25, 37, and 45°C. For salt concentration, NB containing 15% NaCl concentration was diluted using sterile NB to have a range of salt concentrations, i.e. 15, 12.5, 10, 7.5, 5, 2.5, and 1%. The un-inoculated nutrient broth was kept as a control for both experiments. The optimum growth was evaluated by measuring the absorbance at 600 nm in an Epoch 2 microplate spectrophotometer (BioTek Instruments Inc., USA). The experiments were performed in triplicates.
Extraction of the pigments
The pigments were extracted from the cultures by using the solvent extraction method (Numan et al., 2018). The cultures were grown in NB for 7 days at 37°C. After culture growth and visible pigment production, the cells were harvested by centrifugation (4000 rpm for 10 min), re-suspended in 30 ml methanol, and heated in a water bath at 60°C for 30 min. The solutions were centrifuged again at 6000 rpm for 10 min; the supernatants were filtered through a Whatman no. 1 filter paper, collected in Petri plates, and kept at 37°C for methanol to evaporate. The dried pigments were collected and dissolved in 5% methanol for further experiments.
Characterization of the pigments
UV spectral and FTIR analysis
The UV-Vis absorption range of pigments was determined by performing their spectral analysis using UV-Vis spectrophotometry (Epoch 2 microplate spectro-photometer). The pigments solubilized in 5% methanol were analyzed using methanol as blank. The chemical environments of the extracted pigments were determined by FTIR spectroscopy (FTIR 8700, Shimadzu Co., Tokyo, Japan). The samples (10 mg) were mixed homogenously with dried crystals of KBr (200 mg) to make pellets and immediately used for the FTIR analysis (in the range of 4000 to 400 cm−1) to avoid any water absorption.
Thin Layer Chromatography
Spots (a drop made using a glass capillary tube) of the extracted pigment samples (dissolved in 5% methanol) were loaded on commercially available thin layer chromatography (TLC) sheets (stationary phase) (TLC Silica gel 60 F254, MERCK) and run with methanol as a mobile phase. The plate was kept for approximately 20 min at room temperature in the TLC chamber, followed by drying, and then placed in an iodine chamber for developing the spots.
Estimation of the polyphenol content in the pigments
The polyphenol content was determined using the Folin-Ciocâlteu colorimetric method (Sarker et al., 2020c). Tannic acid (0–100 μg/ml in distilled water) was used to prepare the standard curve. The polyphenol concentration in the extracted pigments was quantified in a 96-well plate. Different concentrations of tannic acid (100 μl), 50 μl of Folin-Ciocâlteu reagent, and 50 μl of saturated sodium bicarbonate were added to the wells of a 96-well plate and incubated at room temperature for 1 h. The same protocol was followed for the pigments tested, and 100 μl of 10 mg/ml pigments (in 5% methanol) were used for the same. The absorbance was measured at 760 nm by using an Epoch 2 microplate spectrophotometer. The concentration of polyphenols in the pigments was determined using the equation of the standard graph (Sarker et al., 2020c). The polyphenol content was expressed as micrograms of tannic acid (TA) equivalent. The samples were assayed in triplicates.
Determination of the SPF factor of the pigments
The sun protection factor (SPF) of the pigments was evaluated using a previously reported method (Mansur et al., 1986; Dutra et al., 2004). Briefly, 1 mg/ml pigment in 5% methanol was added in a quartz cuvette, and the absorption spectrum was measured between 290 and 320 nm at 5 nm interval using an Epoch 2 microplate spectrophotometer. Methanol was used as a blank, and the experiment was performed in three replicates. The SPF factor for each pigment was evaluated using the Mansur equation:
where CF represents the correction factor (= 10), I represents the solar intensity spectrum, Abs is the absorbance of the sunscreen product, and EE represents the erythemal effect spectrum. EE × I values are constants (Dutra et al., 2004).
Determination of the antioxidant potential of the pigments
DPPH assay
The ability of the pigments to scavenge free radicals was assessed by the DPPH (1,1-diphenyl-2-picrylhydrazyl) assay (Sarker and Oba, 2019b). For the assay, a stock solution of 150 μl of DPPH (in 0.1 mM in methanol) and 50 μl of 10 mg/ml extracted pigments (in methanol) were used. The reaction mixture was incubated at room temperature in dark for 30 min, and the absorbance was measured at 517 nm in an Epoch 2 microplate spectrophotometer. The positive control used for this experiment was ascorbate (200 μg/ml), and the negative control used was 5% methanol. All samples were assayed in triplicates twice. The formula used to calculate the DPPH scavenging effect was as follows:
ABTS assay
The free radical scavenging ability of the pigments was also evaluated using a more sensitive method, namely the ABTS (2,2N-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) assay (Sarker et al., 2018). A stock solution of 7 mM ABTS and 2.45 mM potassium persulfate was prepared (in distilled water) and mixed in equal volume. The solution was incubated in dark at room temperature for 16 h for radical generation. The blue/green solution of ABTS was then diluted to an absorbance of 0.70 ± 0.02 at 734 nm by adding absolute ethanol. The positive control used for this experiment was ascorbate (200 μg/ml), and the negative control used was 5% methanol. In a 96-well plate, 150 μl of ABTS solution was added to 50 μl of 5 mg/ml pigments (in methanol) and incubated for 30 min at room temperature in dark. Absorbance was determined at 734 nm in an Epoch 2 microplate spectrophotometer. All samples were assayed in triplicates. The percent of antioxidant activity was calculated as follows:
FRAP assay
Ferric reducing antioxidant power (FRAP) of the pigments was evaluated using a previously described method (Pulido et al., 2000). Acetate buffer pH 3.6 (300 mM), FeCl3 6H2O (20 mM), and TPTZ (10 mM in 40 mM HCl) were used as stock solutions. Acetate buffer (25 ml) with TPTZ (2.5 ml) and FeCl3 (2.5 ml) were used as a working solution. The reaction setup used was as follows: 180 μl FRAP reagent, 18 μl D/W, and 6 μl test sample. A standard curve was constructed using FeSO4 7H2O in the range of 100–1000 μmol/l. Ascorbate (62.5 μg/ml) was used as a positive control, and methanol was used as a negative control. The readings were measured at 595 nm using an Epoch 2 microplate spectrophotometer, and the FRAP activity was determined using the equation of the standard graph (y = mx + c ) and represented as μmol Fe2+ equivalent. This experiment was performed in three replicates.
In vitro protective effect of the pigments against H2O2 stress in macrophages
The protective effect of the pigments in the presence of H2O2 stress was shown using the MTT assay (Liu et al., 2010). Murine macrophage cells (RAW264.7) were seeded in a 96-well plate (2 × 104 cells/well) in DMEM + 10% FBS and were allowed to grow at 37°C, 5% CO2 for 24 h in a humidified incubator. After 24 h, the cells were treated with pigments alone or pigments + H2O2 for 24 h. Pigments at a concentration of 500 μg/ml and H2O2 (1 mM) were used for the experiment. After the treatment, MTT (0.5 mg/ml in DMEM) was added to the cells, and the solution was incubated at 37°C for 3 h. The formazan crystals obtained were dissolved with 100 μl of 10% SDS for 2 h. The absorbance was measured at 570 nm using an Epoch 2 microplate spectrophotometer, and the cell survival percentage with respect to control was determined. This experiment was performed in four replicates.
Statistical analysis
All the data from the experiments are expressed as mean ± standard deviation for at least three replicates. The results for “In vitro protective effect of the pigments in macrophages” were analyzed using one-way Analysis of Variance (ANOVA) followed by a posthoc Tukey’s test in GraphPad Prism 8 software. The differences were considered to be significant at P < 0.05.
Results and discussion
Isolation of the bacterial cultures from mangrove soil and water samples, and extraction of the pigments from isolated bacteria
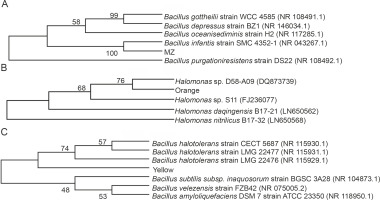
Different colored bacterial colonies were isolated on nutrient agar from mangrove water and soil samples. The bacterial cultures producing pink (MZ) and orange pigment were isolated from mangrove soil, while bacterial culture with yellow pigment was obtained from mangrove water. The isolated strains were identified according to their morphological characteristics and by 16S rDNA sequencing, and the closest prokaryotic matches were determined using the NCBI BLASTN (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Gupta et al., 2015) and the Ribosomal Database Project 11 (http://rdp.cme.msu.edu/) (Khaneja et al., 2009). The sequence alignment was performed using Clustal W (https://www.ebi.ac.uk/Tools/msa/clustalo/), and the phylogenetic tree was assembled using MEGA X (https://www.megasoftware.net/). The colony characteristics, Gram’s staining nature, and the image of the pigment producers are provided in Supplementary Figure 1 and Supplementary Table 1. The 16S rDNA sequencing revealed that the bacterial strain producing pink pigment (MZ) showed 98.84% similarity (NCBI BLAST) and was clustered closely with Bacillus infantis; hence, we concluded that this strain was Bacillus infantis. The bacterial strain producing Yellow pigment showed 73.26% (NCBI BLAST) similarity with different species of Bacillus, indicating that it belonged to Bacillus spp. For the Orange pigment-producing bacterial strain, NCBI BLAST did not yield any matches. However, based on the Ribosomal Database Project 11 (http://rdp.cme.msu.edu/) (Khaneja et al., 2009) and morphological characteristics (Tang et al., 2017), it appeared to be Halomonas spp. Pigment-producing Bacillus (Khaneja et al., 2009; Saha et al., 2017) and Halomonas (Cabrera et al., 2007; Fariq et al., 2019) strains are known to be a part of the soil microflora and are recognized to produce the respective colored pigments. The phylogenetic tree constructed using MEGA X software is shown in Figure 1.
Fig. 1
Phylogenetic relationship tree for A) Bacillus infantis (MZ), B) Halomonas spp. (Orange), and C) Bacillus spp. (Yellow) reconstructed by the neighbour joining method using MEGA X software; the bootstrap value (%) (1000 subsets) indicates the significance of the branches; GenBank accession numbers are given in brackets
Bacteria are a preferred source of pigments because of the controlled and predictable yield (Usman et al., 2017). Here, the pigments were extracted in methanol, and the maximum yield was 0.24–0.26 g/l for all the cultures. This yield is considerably high, and it conforms to the yield reported by Fariq et al. (2019) where the maximum yield of carotenoid pigments in methanol extract was around 0.3 g/l.
Optimization of growth conditions for the bacterial isolates
For large-scale production of the pigments, the incubation temperature and salt concentrations in the media were optimized (Silva-Castro et al., 2019). All the cultures showed higher growth at 1 and 2.5% salt concentration in the medium with OD600 of 0.790 for B. infantis (MZ) and 1.383 for Bacillus spp. (Yellow) at 1% salt concentration, and an OD600 of 0.512 for Halomonas spp. (Orange) at 2.5% salt concentration (Fig. 2A). Our results are in line with those of earlier studies, where carotenoid-producing bacteria were shown to grow at salt concentrations between 0.5% to 14% (Saha et al., 2017), with an optimum growth at 2% NaCl concentration (Kim et al., 2020). Our results also indicate that all the tested cultures can tolerate well up to 15% salt concentrations; however, the bacterial growth decreased at salt concentrations higher than 2.5% (Fig. 2A), and the maximum amount of pigment was produced at 2.5% salt concentration.
Fig. 2
Optimization of growth conditions for the bacterial isolates Bacillus infantis (MZ), Halomonas spp. (Orange), and Bacillus spp. (Yellow) with respect to A) salt concentration and B) temperature
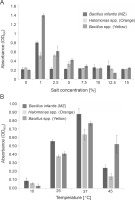
Previous studies have indicated that the optimum temperature for growth and pigment production of different pigment producers was 30–37°C (Khanafari et al., 2010; Gallardo et al., 2016; Chadni et al., 2017). In fact, for various pigment-producing Bacillus and Halomonas species, the optimum temperature for growth and pigment production was found to be 37°C (Cabrera et al., 2007; Saha et al., 2017; Fariq et al., 2019), and our results were similar to the previously reported ones. In our study, all the cultures showed higher growth at 37°C with OD600 of 0.870 for B. infantis (MZ), 0.630 for Halomonas spp. (Orange), and 0.770 for Bacillus spp. (Yellow) (Fig. 2B). The growth of B. infantis (MZ) (OD600 = 0.240) and Halomonas spp. (Orange) (OD600 = 0.140) was reduced at 45°C, but the high temperature did not affect the growth of Bacillus spp. (Yellow) (OD600 = 0.520) (Fig. 2B). These results also indicate that all the three cultures could survive in a wide range of temperatures, i.e., between 10–45°C; however, the optimum temperature for the highest pigment production was found to be 37°C.
The survival of the tested bacterial isolates in a wide range of salt concentrations and temperatures could be attributed to their habitat and pigment production, as the production of pigments helps bacteria to easily adapt to their respective environments (Sutthiwong et al., 2014; Ramesh et al., 2019). Mangrove rhizosphere is known to have salinity and temperature fluctuations (Simőes et al., 2015; Kharangate-Lad and Bhosle, 2016), and it is evident that these bacteria have adapted to their natural habitat.
Characterization of the pigments
Partial characterization of the pigments was performed by UV-Vis spectroscopy and FTIR analysis. TLC was performed to assess the components present in the crude pigment extracts (Mulik et al., 2017) (Supplementary Fig. 2). In UV-Vis analysis, the pigments showed absorbance in both UV and visible range, indicating that they had very broad absorption spectra (Fig. 3A). Carotenoids are known to absorb in the UV range (Aust et al., 2005), and carotenoids extracted from extremophilic bacteria that are constantly exposed to solar radiation are also known to protect against UV-A (400–315 nm), UV-B (315–280 nm), and UV-C radiation (280–100 nm) (Mohana et al., 2013; Reis-Mansur et al., 2019). In our study, the absorption maxima for MZ, Orange, and Yellow pigments in the UV range were 230 nm, 210 nm, and 250 nm, respectively (Fig. 3A); this indicated that these pigments could be carotenoids. The absorption maxima in the visible range for these pigments were 480 nm, 460 nm, and 410 nm for MZ, Orange, and Yellow pigments, respectively (Fig. 3A). According to the data reported in the literature, the characteristic absorbance peaks of carotenoids are between 400 nm and 500 nm (Scott, 2001; Khaneja et al., 2009; Zaghdoudi et al., 2017; Silva-Castro et al., 2019). The visible ranges of absorption for the extracted pigments were between 410 nm and 480 nm, which is similar to those described earlier and therefore indicate that these pigments were carotenoids.
Fig. 3
Characterization of pigments: A) UV-Vis spectrum of pigments indicating that all the three pigments (MZ, Orange, and Yellow) absorb in both UV and visible range and B) the FTIR spectra of the pigments

In the FTIR analysis, the three pigments showed spectrum patterns similar to those reported for carotenoid pigments (Usman et al., 2018; Cojoc et al., 2019) and derivatives of beta-carotene structure (Neha et al., 2017). The peaks at 1395 cm−1 and 1449 cm−1 represented C-H bending frequency along with the presence of methyl groups, and the peak at 2915 cm−1 represented C−H stretching (Cojoc et al., 2019). Additionally, peaks were also observed at 1620 cm−1, which indicated the presence of conjugated double bonds of the carbonyl functional group (Usman et al., 2018). The broad peak between 3300 and 3500 cm−1 indicated the presence of the OH or NH group (Cojoc et al., 2019) (Fig. 3B).
Collectively, on the basis of the results of UV-Vis spectra and FTIR analysis, it was concluded that these extracted pigments were carotenoids.
Estimation of polyphenol content in the pigments and determination of its SPF
Phenolic compounds are the most studied phytochemicals and are also the major contributors to the antioxidant activity of herbal plants and spices (Shahidi and Ambigaipalan, 2015; Martins et al., 2016). Although plants are the major source of polyphenols, some evidence shows the production of phenolic compounds by microorganisms as secondary metabolites (Kawahara et al., 2012; Bhanja Dey et al., 2016; Tan et al., 2018). Therefore, the polyphenol content of the pigments under this study was estimated using the Folin-Ciocâlteu assay and was found to be the highest for Orange pigment (48.8125 μg TA equivalent), followed by Yellow (46.5625 μg TA equivalent) and MZ (22.25 μg TA equivalent) (Fig. 4A). In previous studies, the pigments extracted from microorganisms containing polyphenols have been shown to exhibit antioxidant activities (Pawar et al., 2015; Gonçalves Tavares et al., 2018), thus indicating an association of polyphenols with pigments, which contributes to the antioxidant activity of the pigments.
Fig. 4
Estimation of A) polyphenol content in the pigments using the Folin-Ciocâlteu method and B) sun protection factor (SPF) values of the pigments

Fig. 5
Antioxidant activity of MZ, Orange, and Yellow pigments demonstrated through A) DPPH (2,2-diphenyl-1-picrylhydrazyl) assay, B) ABTS (2,2-azinobis-3-ethyl-enzothiazoline-6-sulfonicacid) assay, and C) FRAP (Ferric reducing antioxidant power) assay, indicating that these pigments have high antioxidant potential
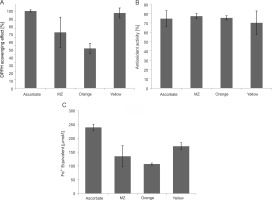
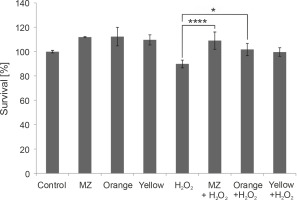
Fig. 6
In vitro protective effect of the pigments against H2O2 stress in macrophage cell line RAW264.7, indicating that MZ and Orange pigment significantly protect macrophages from H2O2-induced cell death. (****P < 0.0001, *P = 0.0212)
The absorption of the pigments in the UV range indicated that these pigments could act as UV-protective agents. This characteristic was exploited further for the determination of the SPF values of the pigments by using the Mansur equation (Dutra et al., 2004). It was observed that Orange showed the highest SPF value as compared to the other two pigments. The SPF values were 5.22, 4.71, and 3.99 for Orange, Yellow, and MZ, respectively (Fig. 4B). The values obtained in this study were found to be higher than those reported for the pigments of Exiguobacterium sp. and Serratia marcescens, where the SPF values were found to be in the range of 1.6 to 2.72 (Bana and Heshmatipour, 2014). The effective role of carotenoids in protection against UV light has been reported earlier (Reis-Mansur et al., 2019). It has also been shown that the SPF values of commercial sunscreens can be increased by adding bacterial or fungal pigments (Suryawanshi et al., 2015; Koli et al., 2019). The SPF value of commercial sunscreens increased by 10–22, 20–65, 36.5, and 13% following the addition of violacein, prodigiosin (Suryawanshi et al., 2015), red pigment, and yellow pigment produced by Monsacus purpureus (Koli et al., 2019), respectively. Because the pigments tested in the present study also have high SPF values, they have the potential to be incorporated into the sunscreens to increase their UV protectiveness.
Determination of the antioxidant potential of the pigments
A single assay is not accurate enough in determining the antioxidant potential for all the groups of antioxidant compounds; therefore, different assays should be performed to facilitate better comparison and for drawing conclusions (Barros et al., 2017). Hence, in the present study, three assays were performed. DPPH and ABTS assays determine the potential of the pigments to scavenge free radicals (Mohamed et al., 2016). The percent of radical scavenging activity was calculated for the pigments, and all the studied pigments showed significant DPPH scavenging activity. Yellow showed the highest antioxidant activity (88.62%) comparable with the ascorbic acid standard (90.95%), followed by MZ (65.81%) and Orange (46.8%) (Fig. 5A). Bacterial carotenoids were previously reported for their antioxidant activities (Manimala and Murugesan, 2014). Approximately 60–80% antioxidant activity through the DPPH assay was previously reported for pigments extracted from bacteria isolated from mangrove environments (Umamaheswari and Indra, 2013; Raghava Rao and Raghava Rao, 2013; Subramanian and Gurunathan, 2020). In the present study, the extracted pigments exhibited similar antioxidant activity, with the highest antioxidant activity of 88.62% shown by Yellow pigment.
The studied pigments were also able to scavenge ABTS radicals. MZ (77.26%) and Orange (75.37%) showed the highest antioxidant activity comparable to the ascorbic acid standard (74.56%), followed by Yellow (70.09%) (Fig. 5B). Our results were similar to those of a previous study where carotenoid pigments from Meiothermus sp. were reported to have an ABTS scavenging activity of around 74.3% (Mukherjee et al., 2017). However, the pigments in our study showed much higher values than those reported by Park et al. (2018), where carotenoids from Arthrospira platensis showed 33.7% antioxidant activity in the ABTS assay.
FRAP assay also evaluates the antioxidant activity of the pigments (Rizk et al., 2014). By observing the reduction of the colorless Fe3+-TPTZ to indigo-colored Fe2+-TPTZ, the total reducing capacity of the samples was measured, and the antioxidant potential was calculated (Rizk et al., 2014). Among the three pigments, the FRAP value was the highest for Yellow pigment (171.39 μmol Fe2+ equivalent), followed by MZ (135.08 μmol Fe2+ equivalent) and Orange (106.75 μmol Fe2+ equivalent) (Fig. 5C). The FRAP value of the ascorbic acid standard was 239.65 :mol Fe2+ equivalent (Fig. 5C). Our results are in line with previous studies, where significant FRAP activity exhibited by pigments was reported (Salem et al., 2014; Pawar et al., 2015).
Collectively, our data indicated that all three pigments are potent antioxidants. Yellow pigment showed the highest antioxidant potential as compared to the other two pigments in DPPH and FRAP assays. However, in the ABTS assay, MZ showed the highest antioxidant potential. Flavonoid compounds are reported to be more sensitive toward the DPPH assay than toward the ABTS assay (Mohamed et al., 2016); this indicates that Yellow may be a mixture of flavonoids and carotenoids, as observed in other natural products (Wan et al., 2019).
In vitro protective effect of the pigments against H2O2-induced stress in macrophages
Because the pigments showed antioxidant properties in chemical assays, their ability to act as antioxidants in the presence of H2O2 was studied by the MTT assay in murine macrophages. Mouse macrophages (RAW264.7) are known to have a high amount of reactive oxygen species (ROS) and hence act as a good model to study the antioxidant characteristics of any naturally extracted compounds (Castaneda et al., 2017). Exposure to H2O2 triggers oxidative stress conditions and cell death (Kwon et al., 2019). Previous studies have indicated that pigments prevent H2O2-induced cell cytotoxicity in retinal pigmented epithelial cells (Huang et al., 2018), HepG2 liver cancer cell line (Fang et al., 2018), and RAW264.7 murine macrophage cell line (Wang et al., 2019). Here, we performed a similar study in macrophages. When macrophages were subjected to H2O2 stress, MZ and Orange pigment showed significantly higher protection by increasing the survival percent to 109.016 and 101.7%, respectively, as compared to 1 mM H2O2 control, which was 89.9%. Yellow also showed a protective effect against H2O2 stress; however, it was not as effective as MZ and Orange (Fig. 6).
In previous studies, pigments such as anthocyanins have been shown to prevent H2O2-induced cytotoxicity in human retinal pigment epithelial cells, wherein blueberry anthocyanin extract (BAE) and malvidin were found to increase the percentage of viability from 63.69% in H2O2-treated cells to 86.57% in BAE-treated cells and to 115.72% in malvidin-treated cells (Huang et al., 2018). Anthocyanins were also reported to rescue H2O2-damaged liver cancer cell line HepG2, wherein the red cabbage anthocyanin extract was found to considerably increase the percentage of viability of H2O2-treated HepG2 cells (Fang et al., 2018). In one study, astaxanthin, which is one of the xanthophyll carotenoids, was shown to increase the viability of H2O2--treated RAW264.7 macrophage cells by 49.4% (Wang et al., 2019).
Our results strongly agree with the results of the abovementioned studies. The results of our experiments were also comparable to the restorative effect of the known antioxidant compounds, namely glutathione (Kwon et al., 2019) and ascorbate (Oh et al., 2020), wherein glutathione was shown to protect RAW264.7 murine macrophages cells and ascorbate was shown to protect ARPE-19 cells (retinal pigment epithelial cells) from H2O2-induced cell death by significantly increasing the cell viability of H2O2-damaged cells (Kwon et al., 2019; Oh et al., 2020). These findings indicate that the pigments can protect cells from H2O2-induced cytotoxicity and act as potent antioxidants.
Conclusions
In the present study, three different colored pigments were extracted from bacteria isolated from the mangrove rhizosphere habitat. The pigments absorbed in the UV and visible range and had a good SPF value, indicating their potential UV protective properties. The extracted crude pigments had high polyphenol content. Chemical assays and an in vitro cell line-based assay showed that these pigments have high antioxidant potential. Because synthetic antioxidants are known to have adverse side effects, natural and more potent antioxidants will be preferred in future. These natural pigments can be developed as a safer and more effective alternative to the currently used synthetic antioxidants.










