Introduction
Crop fields worldwide are facing the crisis of phosphorus (P) deficiency. P deficiency is the major cause of decline in crop yield in nearly 30% of agricultural lands; it impedes the normal growth of plants, thus acting as a limiting factor that affects plant growth after nitrogen and potassium (Carstensen et al., 2018). Despite high chemical inputs, soils globally are becoming depleted in P content critically if not solely, due to inefficient organic P management (Alewell et al., 2020). The solution to this issue is the so-called “legacy” P known as P reserves. P reserves in the soil occur exclusively in the form of organic and inorganic P reserves. The inorganic P reserved pool comes from the weathering of minerals and excessive fertilizer input that gets deposited in soil as hydrate oxides of calcium, aluminum, manganese, and iron. Organic P exists in the form of sugar phosphates, inositols, nucleic acids, phospholipids, nucleotides, phytate, or phytin that gets deposited in soil from plant residues and manures (Hata et al., 2010) and constitutes up to 30–80% of the total P reserves (Cao et al., 2018).
The plant growth-promoting rhizobacteria (PGPR)-based biofertilizer technology is viewed as a reasonable alternative to revitalize this P pool from soil for plant use as phospholipids and phytate can be hydrolyzed by the microbial enzymes phosphatase and phytases (Irshad and Yergeau, 2018). Phytate-mineralizing bacteria (PMB) catalyze the conversion of insoluble P into a soluble form that can be assimilated by plants. The solubilization of inorganic forms of P, such as rock phosphate, hydroxyapatite, dicalcium phosphate, and tricalcium phosphate, by microbial activity has been well reported in the literature (Kaur and Kaur, 2020).
The plant growth-promoting potential of rhizospheric bacteria capable of P solubilization is also well elaborated, thus depicting their potential for use as biofertilizers. Studies related to PMB isolated from rhizospheric soils of Punjab are, however, poorly reported. Only a few studies have reported on rhizospheric bacteria with an ability to solubilize and mineralize both inorganic and organic P (Sharma et al., 2013). In the present study, PMB were isolated from rhizospheric soils of Punjab, and their plant growth-promoting potential was determined. It will be intriguing to know the plant growth-promoting potential of PMB, so that PMB-based biofertilizers can be formulated for indigenous crops.
Materials and methods
Isolation of PMB from rhizospheric soil
PMB were isolated using Pikovskaya’s medium containing D-glucose (10 g/l), calcium chloride (2 g/l), ammonium nitrate (5 g/l), potassium chloride (0.5 g/l), ferrous sulfate (0.01 g/l), magnesium sulfate (0.5 g/l), manganese sulfate (0.01 g/l), phytic acid sodium salt hydrate (5 g/l), and agar (15 g/l) (Hosseinkhani et al., 2009). Soil samples from the rhizospheric niches were collected from sub-mountainous regions of Mohali (30°53′N, 76°38′E) and Gurdaspur (32°2′30.9948″N, 75°24′19.2024″E) and the central districts Nawanshahr (31.1167°N, 76.1333°E) and Ludhiana (30.9010°N, 75.8573°E) of Punjab. Soil (1 g) was homogenized in 9 ml of distilled water and serially diluted up to 10–9 dilutions. Each aliquot was spread plated on Pikovskaya’s agar and incubated at 28°C. The colonies showing clear zones after 48–168 h of incubation were selected and purified. The solubilization index (SI) was calculated by measuring the diameter of the colony and the diameter of the halo zone after 48, 96, and 168 h of incubation by using the formula:
Soluble P formed by the microbial action was quantified using the method of Bray and Kurtz (1945). Pikovskaya’s medium supplemented with phytic acid sodium salt hydrate was inoculated with the microbial culture at the density of 12 × 108 CFU/ml and allowed to grow at 28°C. To analyze the soluble P formed, 2 ml of sample was taken aseptically at 48, 96, and 196 h. The supernatant was obtained by centrifuging the sample at 11410 g for 10 min, and the amount of soluble P was determined by the phosphorus-molybdenum blue colorimetric method (Bray and Kurtz, 1945).
Biochemical characterization and genetic identification of bacteria
The isolates were biochemically characterized using the methods described in Bergey’s Manual (Cappuccino and Shermann, 1992). The best isolates showing high plant growth-promoting potential were identified by the 16S ribosomal DNA (rDNA) sequencing method. For this purpose, the bacterial culture at optical density of 0.5 was used for genomic DNA extraction (Pamidimarri et al., 2009). The extracted genomic DNA was amplified with a 16S forward primer (5′–GGATGAGCCCGCGGCCTA–3′) and a 16S reverse primer (5′–CGGTGTGTACAA GGCCCGG–3′) using PCR polymerase (Heuer et al., 1997). After 35 PCR cycles consisting of a single step of initial denaturation for 3 min, followed by repeated cycles of denaturation for 1 min at 94°C, annealing for 1 min at 55°C, and extension for 2 min at 72°C, the final extension was performed at 72°C for 7 min. Gel electrophoresis was used to purify the PCR product, which was then sequenced using the Big Dye Terminator sequencing machine version 3.1 (ABI 3130 Genetic Analyzer). The sequences were further aligned with reference 16S rDNA sequences of Burkholderia sp. from the NCBI database using the BLAST program and analyzed to identify the bacteria and its closest neighbors by using MEGA7 software (Kumar et al., 2016).
Tricalcium phosphate solubilization assay
The ability of the bacterial isolates to solubilize inorganic source of P, namely tricalcium phosphate (TCP), was determined by inoculating 10 μl of fresh bacterial culture of 0.5 McFarland standard on Pikovskaya’s agar plates and allowed to grow at 28°C for 168 h. The isolates were observed for the appearance of clear zones around the colonies, and the solubilization index was measured after 48, 96, and 168 h of incubation as described above. An in vitro quantitative assay to quantify soluble P formed was performed by inoculating the fresh culture in Pikovskaya’s broth supplemented with TCP according to the method described by Bray and Kurtz (1945), as mentioned above.
Acid phosphatase activity
The bacterial cultures were inoculated in Pikovskaya’s broth supplemented with TCP and incubated at 28°C. Following the incubation period, the bacterial culture was aseptically collected and centrifuged at 11 410 g for 10 min at 4°C. The supernatant obtained (0.3 ml) was added to a reaction mixture of p-nitrophenyl phosphate (1.2 ml) and incubated at 30°C for 1 h. The reaction was stopped by adding 1 N NaOH (3 ml). The p-nitrophenol formed by the action of acid phosphatase on p-nitrophenyl phosphate was measured at 410 nm by using a spectrophotometer. Enzyme activity was expressed as micromoles of p-nitrophenol formed per hour per milliliter (μM of p-nitrophenol/ml/min) of culture supernatant by using the standard graph plotted with p-nitrophenol (Dorn and Rivera, 1966).
Alkaline phosphatase activity
Acid phosphatase production by the isolates was determined by growing the bacterial cultures in TCP-enriched Pikovskaya’s broth. The bacterial culture was aseptically drawn at 48, 96, and 196 h of incubation and centrifuged for 15 min at 4°C at 11 410 g to remove bacterial cells. The supernatant (1 ml) was mixed with 4 ml of modified universal buffer (pH 11.0), followed by the addition of 1 ml of 0.025 mM disodium p-nitrophenyl phosphate. The mixture was incubated at 37°C for 1 h. The reaction was stopped by adding 4 ml of 0.5M NaOH and 1 ml of 0.5M calcium chloride. The contents were filtered through a Whatman No. 42 filter paper. The concentration of p-nitrophenol was measured at 420 nm by using a UV-vis spectrophotometer (Tabatabai and Bremner, 1969). Enzyme activity was expressed as micromoles of p-nitrophenol released per hour per milliliter (μM of p-nitrophenol/ml/min) of culture filtrate using the standard graph plotted with p-nitrophenol.
Phytase activity
The phytase activity of the isolates was determined by inoculating fresh bacterial cultures in Pikovskaya’s broth containing phytic acid sodium salt hydrate (Sigma-Aldrich, USA) and incubating at 28°C for 24 to 196 h. Cell-free supernatant was obtained by centrifuging the bacterial culture at 11 410 g for 15 min at 4°C to harvest bacterial cells. The supernatant (0.2 ml) was incubated in water bath at 37°C for 5 min, and 0.2 ml of 1% (w/v) sodium phytate solution was added. After 15 min, 0.4 ml of 15% trichloroacetic acid was added. The mixture obtained was centrifuged at 1008 g for 10 min, and the supernatant (0.2 ml) was transferred to another tube containing 1.8 ml of MilliQ water (Millipore). A freshly prepared color reagent (2 ml) was added to the mixture and incubated at 50°C for 15 min. The absorbance was read at 820 nm against a water blank. The results were interpreted using the graph of standard KH2PO4 solutions at different concentrations, and the enzyme activity was expressed as micromoles of phosphate released per hour per milliliter of culture material (Kim and Lei, 2005).
Plant growth-promoting attributes
Various plant growth-promoting attributes of the isolates were studied according to the standard procedures reported in the literature. Indole acetic acid (IAA) and gibberellic acid (GA) production was detected by the method of Bent et al. (2001) and Borrow et al. (1955), respectively. Ammonia production was determined using the standard method of Cappuccino and Sherman (1992). Potassium solubilization was estimated by growing the bacterial isolates on an Aleksandrov agar medium (Aleksandrov et al., 1976). Nitrogen-fixing ability of the isolates was tested by growing them on an Nfb semi-solid medium as described by Baldani et al. (2014). Zinc solubilization was determined by growing the isolates on Bunt and Rovira medium containing 0.1% zinc oxide as the insoluble form of zinc (Bunt and Rovira, 1955). Siderophore production was analyzed using a modified microplate procedure as described by Arora and Verma (2017). Siderophore produced was measured in percent siderophore unit (psu) by using the formula:
where Ar is the absorbance of reference and As is the absorbance of sample.
Cellulase hydrolysis test, catalase test, protease production, and hydrogen sulfide production test for the isolates were performed using the methods defined in Bergey’s Manual of Systematic Bacteriology (Cappuccino and Sherman, 1992). ACC (1-aminocyclopropane-1-carboxylate) deaminase production was tested by growing the isolates on a minimal DF (Dworkin and Foster) salts medium supplemented with ACC (Dworkin and Foster, 1958).
Statistical analysis
The data are presented as mean ± SD of three replicates. The equal distribution method was used to categorize the isolates into low, medium, and high categories by subtracting the minimum value from the maximum value, and the difference between the two was divided by 3 to obtain the three classes with an equal interval. The mean values were subjected to Tukey’s test by using XLSTAT 2021. The dendrogram was prepared by clustering bacterial isolates using XLSTAT 2021.
Results and discussion
Screening of the isolates for phytate-mineralizing ability
From each district with a geographically diverse location, four soil samples were collected. Each sample was collected in triplicates and mixed to form a composite sample. From the 16 soil samples, the total bacterial colonies ranging from 1.84 × 105 to 6.20 × 105 colony forming unit (CFU) were found, of which 34 bacterial isolates showed potential clear zones around the colonies when grown on a Pikovskaya’s agar medium supplemented with phytic acid sodium salt hydrate. Only 2.35% of the total bacterial colonies showed phytate-mineralizing ability. Previous studies by Tao et al. (2008) have shown significantly lower proportion of PMB (0.205%) in rhizospheric soils. The isolates were repeatedly subcultured to confirm any loss of phytate-mineralizing activity, and the solubilization index was measured after 48, 96, and 196 h of incubation. The maximum SI was observed for isolate A33 (11.23) after 48 h of incubation, which decreased on 96 h of incubation. However, for most of the other isolates, the SI increased up to 96 h of incubation. Seven isolates exhibited the maximum SI after 48 h of incubation, 16 isolates showed maximum solubilization after 96 h of incubation, while 11 isolates showed the maximum SI after 196 h of incubation. Significant differences in SI were found within the isolates. Isolates A32, A27, and A28 showed high SI varying from 7.81 to 9.56, 11 isolates exhibited medium SI ranging from 6.06 to 7.81 while 20 isolates showed low SI varying from 4.31 to 6.06 (Table 1). Earlier studies have also shown a decrease in SI by phosphate-solubilizing bacteria (PSB) after 72–96 h of incubation (Chen and Liu, 2019).
Table 1
Phytate mineralization, phosphate solubilization, and related enzyme activities of the isolates at 96 h of incubation
Biochemical identification revealed a wide range of PMB (Supplementary Table 1). The isolates belonged to Pseudomonas, Enterobacteriaceae, Bacillaceae, Paenibacillaceae, Micrococcaceae, Burkholderiaceae, Flavobacteriaceae, and Streptococcaceae families. Previous studies have reported that bacterial species belonging to these families possess potential P solubilizing abilities (Kaur and Kaur, 2020; Karnwal, 2021). These bacteria were also shown to have promising plant growth-promoting traits.
Quantification studies were performed to determine the efficiency of bacterial isolates to solubilize sodium phytate. The amount of soluble P formed was observed to be the highest at 96 h of incubation, which decreased at 168 h of incubation. The decrease in soluble P can be attributed to the high amount of P needed to sustain bacterial growth or due to secondary solubilization that causes reprecipitation of the phosphate compound into brushite (Goenadi and Sugiarto, 2000). Bacterial isolates vary significantly in their potential to mineralize sodium phytate. The equal distribution method was used to categorize the isolates into high, medium, and low P solubilizers. Eleven isolates were categorized as high sodium phytate mineralizers, with soluble P formed in the range of 84.62–105.93 μg/ml. Twenty isolates were categorized as medium solubilizers with soluble P formed varying from 63.42 to 84.62 μg/ml. Only three isolates with soluble P production in the range of 42.22–63.42 μg/ml were categorized as low P producers (Table 1). Tao et al. (2008) reported that the bacteria isolated from soils of flooded and non-flooded plains showed very low organic P-mineralizing potential ranging from 8.2 to 17.8 μg/ml. Of 40 bacterial isolates obtained from a banana tree, only 7.5% of the isolates could mineralize organic P (Matos et al., 2016). In the present study, a higher proportion of sodium phytate mineralizers were isolated, indicating that organic P reserves in the soil can be potentially utilized as a source of fertilization through bacteria.
TCP solubilization potential
The isolates were tested for their potential to solubilize an inorganic P source (TCP). For TCP solubilization, potential SI was observed in all the isolates at 48 h of incubation, which increased at 96 h of incubation. However, no further increase in the SI was observed. A significant variation was observed in mean SI of the isolates, ranging from 1.12 to 4.99. Chakkaravarthy et al. (2010) reported decrease in SI after 72–96 h of incubation by phosphate-solubilizing Bacillus sp. isolated from rhizospheric soils of sorghum, groundnut, and cotton from Chennai, India. In our present study, four isolates with SI in the range of 3.7–4.99 were categorized as high TCP solubilizers. Twenty-three isolates exhibiting SI in the range of 2.41–3.7 were medium TCP solubilizers. Seven isolates with SI ≤ 2.41 were categorized in the low category. The content of soluble P formed by the isolates was found to be the maximum at 96 h of incubation and ranged from 3.12 to 110.88 μg/ml. The equal distribution method was used to categorize the bacterial isolates into low, medium, and high P solubilizers, which depicted H ≥ 74.96 and L ≤ 39.04. Fourteen isolates were high P solubilizers with soluble P content ranging from 74.96 to 110.88 μg/ml, 16 isolates were medium P solubilizers with soluble P content ranging from 39.04 to 74.96 μg/ml, and four isolates produced low soluble P content ranging from 3.12 to 39.04 μg/ml (Table 1). Matos et al. (2016) assessed the TCP solubilization potential of 40 bacterial isolates from the rhizospheric soils of banana and found that 67.5% of the bacterial isolates showed SI within the range of 1.22–4.82. Compared to this previous study, the bacterial isolates in our study showed better P solubilizing potential.
Phytase activity
Phosphate solubilization and mineralization by rhizospheric bacteria is partly due to secretion of enzymes involved in the dephosphorylation of phosphoester and phosphoanhydride bonds. These enzymes are nonspecific phosphomonoesterases and can be either acid or alkaline phosphatases (Jorquera et al., 2011). Both acid and alkaline phosphatases are involved in the solubilization of inorganic phosphate compounds. The phytate mineralizing potential is, however, catalyzed by secretion of the enzyme phytases (Richardson et al., 2001). In the tested isolates, phytase activity was found to be the maximum at 96 h of incubation. All the isolates exhibited phytase activity ranging from 0.01 to 0.97 μM of pi/ml/min. Singh et al. (2014) reported that the phytase activity of a PGPR strain was in the range of 0.076–0.174 U/ml. Our study reports much higher phytase production in rhizospheric bacteria. Nineteen isolates showed high phytase activity, 13 isolates were medium phytase producers, and two isolates were categorized as low phytase producers.
Acid and alkaline phosphatase activity
The isolates obtained in the present study exhibited both acid and alkaline phosphatase activity (Table 1). Significant differences were observed in the acid phosphatase activity among the isolates, ranging from 0.03 to 0.91 μM of p-nitrophenol/ml/min. Seventeen isolates exhibited low acid phosphatase activity of ≤0.32, while seven isolates showed a high acid phosphatase activity of ≥ 0.61. Ten isolates exhibited medium acid phosphatase activity ranging from 0.32 to 0.61 μM of p-nitrophenol/ml/min. Except for isolates A5, A13, and A16, all the isolates showed acid phosphatase activity after 48 h of incubation, and the activity increased at 96 h of incubation. Low enzyme activity was found at 168 h of incubation. A similar trend was noted in alkaline phosphatase activity. The enzyme activity was the highest after 96 h of incubation, varying from 0.03 to 0.89 μM of p-nitrophenol/ml/min. Fifteen isolates showed low alkaline phosphatase activity (0.03–0.316), 13 isolates exhibited medium activity (0.316–0.602), and six isolates showed high alkaline phosphatase activity (0.602–0.88 μM of p-nitrophenol/ml/min). Phosphomonoesterase activity studied in 36 isolates from the rhizospheric soil of Doryanthes excelsa and Pueraria montana ranged from 0.106 ± 0.021 μM of p-nitrophenol/ml/min and 0.086 ± 0.005 μM of p-nitrophenol/ml/min, respectively (Cabugao et al., 2017). The phosphomonoesterase activity in the present study was much higher than that reported previously, indicating that the isolates of the present study have a higher potential for P solubilization due to phosphomonoesterase production.
Plant growth-promoting potential
Rhizospheric bacteria are known to secrete the most native auxin (IAA), which is the key hormone that enables root elongation (Olatunji et al., 2017). Several studies have reported that rhizospheric bacteria benefit the plants through enhanced root elongation, nodule formation, and overall development (Spaepen et al., 2010; Kaur and Kaur, 2021; Swarnalakshmi et al., 2021). Therefore, the IAA production characteristic of the rhizospheric bacteria is a functional trait that determines its plant growth-promoting characteristic. All the tested isolates exhibited IAA production, the concentration of which increased up to 168 h of incubation (Table 2). Significant differences were observed in IAA production among all the isolates, with concentrations varying from 25.90 to 37.54 μg/ml. According to the equal distribution method, the maximum number of isolates showed medium IAA production with concentrations ranging from 37.54 to 49.18 μg/ml. Nine isolates showed IAA production at ≤ 37.54 and ≥ 49.18 and were thus grouped as low and high IAA producer categories, respectively. Lebrazi et al. (2020) studied IAA production in 80 rhizospheric bacteria. Although the majority of the isolates produced IAA, only a few isolates exhibited high IAA concentrations ranging from 105 to 135 μg/ml (Lebrazi et al., 2020). IAA production in the well-characterized plant growth-promoting Enterobacter sp. MS32 and Pseudomonas sp. MS16 was reported to be 28.1 and 25.6 μg/ml, respectively (Suleman et al., 2018). Vikram et al. (2007) reported the amount of IAA produced by 30 different PSB strains in the range of 0.044–1.121 μg/ml. This difference in IAA production may be because the production of IAA varies in different species and in strains of the same species (Özdal et al., 2016). Moreover, several environmental factors also cause differences in the production of IAA (Zerrouk et al., 2019; Jasim et al., 2014).
Table 2
Indole acetic acid production (μg/ml) by the isolates at different periods of incubation
GA is another phytohormone responsible for plant physiology and development, seed germination, and stem and leaf growth (Gupta and Chakrabarty, 2013). Many bacterial species produce GA as a secondary metabolite that acts as a signaling molecule in the host plant (Salazar-Cerezo et al., 2018). In the present study, GA production in the bacterial isolates was observed to increase from 48 to 96 h of incubation and then decreased up to 168 h of incubation, except in isolates A26 and A29 (GA production was maximum after 168 h of incubation in these isolates). Significant differences were observed in the GA concentration among all the isolates, ranging from 12.34 to 100 μg/ml (Table 3). Nine isolates produced a low amount of GA ranging from 12.34–41.56 μg/ml. Four isolates produced a medium amount of GA (41.56–70.78 μg/ml), and the other 21 isolates showed high GA production (≤ 70.78 μg/ml). Saleemi et al. (2017) reported that GA production in rhizospheric bacteria varied from 10.0 to 14.8 mg/l. Plant growth-promoting Pseudomonas strains showed GA production ranging from 25.92 to 43.32 μg/ml (Sharma et al., 2017). The isolates obtained in the present study showed much higher GA production than that reported in the literature, thus suggesting that they have a better PGPR potential.
Table 3
Ammonia, gibberellic acid, and siderophore production of the isolates at 96 h of incubation
Certain rhizospheric bacteria have the potential to augment plant biomass by producing ammonia that can act as a direct source of nitrogen supply to the plants (Marques et al., 2010). In the tested isolates, ammonia production was observed to be maximum at 96 h of incubation and then subsequently decreased significantly up to 168 h of incubation. Significant differences were found in ammonia production among the isolates, ranging from 12.66 to 43.92 μmol/ml. Ten isolates produced a medium amount of ammonia (23.08–33.5 μmol/ml), while 10 isolates produced higher amounts of ammonia (33.5–43.92 μmol/ml) (Table 3). Fourteen isolates produced a low amount of ammonia in the range of 12.66–23.08 μmol/ml. Earlier studies have shown that ammonia production in rhizobacteria ranged from 2.5 to 7.54 μmol/ml (Bhattacharyya et al., 2020). Dutta and Thakur (2017) also reported the range of ammonia produced by rhizobacteria from tea plants in the range of 2.3–6.2 μmol/ml. The isolates obtained in the present study showed potent ammonia-producing ability as compared to that reported in earlier studies. These differences might be due to the differences in the environment from which they are isolated or might be due to genomic differences.
Siderophores are low-molecular-weight compounds with a high affinity for Fe(III) and are produced by various bacteria to sequester iron during low availability. Because iron deficiency is the major factor that limits crop yield, siderophore-producing rhizobacteria offer great benefits by compensating for iron deficit (Ferreira et al., 2019). Earlier studies have shown PSBs produce 80–89.77% siderophores (Kshetri et al., 2018), while other PSBs produce 27.86-95.22% siderophores (Gupta et al., 2012). The Streptomyces roseocinereus MS1B15 strain was reported to produce siderophores up to 14.09 psu (Chouyia et al., 2020). In the present study, 15 isolates produced a low amount of siderophores, varying from 21.01% to 25.84%. Eighteen isolates produced siderophores in a medium amount, ranging from 25.84% to 30.67%. Only one isolate (A30) produced a high amount of siderophores (35.49%) (Table 3).
All the 34 isolates were also screened for nitrogen fixation; zinc and potassium solubilization; and production of hydrogen sulfide, catalase, cellulase, and protease. Of the 34 isolates, 55.88% isolates showed nitrogen fixing activity, 73.53% isolates showed potential zinc solubilization, 47.06% isolates exhibited potential potassium solubilization, and 47.05% isolates showed catalase production. However, only 26.47%, 17.64%, and 23.53% isolates showed hydrogen sulfide, cellulase, and protease production, respectively (Table 4).
Table 4
Screening of isolates for other plant growth-promoting traits
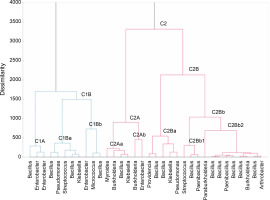
On the basis of differences in phosphate solubilization and various plant growth-promoting traits, clustering of 34 bacterial isolates was performed using XLSTAT 2021 (Fig. 1). The bacterial isolates obtained in this study were divided into two main clusters in the dendrogram, namely C1 and C2. Cluster C1 has 2 subclusters: C1A and C1B. Subcluster C1A includes isolates belonging to the genera Bacillus sp. and Enterobacter sp. These isolates exhibited medium/low phosphate solubilization and phytohormone production and medium/high siderophore and ammonia production. C1B has 2 subclusters: C1Ba and C1Bb. Isolates included in subcluster C1Ba were biochemically identified as Bacillus sp., Streptococcus sp., Klebsiella sp., and Pseudomonas sp.; they exhibited low/medium phosphate solubilization, low/medium phytohormone production, and medium siderophore and ammonia production. In contrast, the isolates of the C1Bb cluster belonging to the genera Enterococcus sp., Micrococcus sp., and Bacillus sp. were low TCP solubilizers and produced phytohormones. Siderophore production was also low, while high ammonia production and high/medium phytate mineralization were observed. Cluster C2 has 2 subclusters, namely C2A and C2B, wherein C2A again has 2 subclusters: C2Aa and C2Ab. C2Aa contains isolates biochemically identified as belonging to the genera Myroides sp., Burkholderia sp., Bacillus sp., and Klebsiella sp.; these isolates exhibited medium/low TCP solubilization and siderophore and ammonia production. Medium/high phytate mineralization and phytohormone production was observed in strains assigned to cluster C2Aa. The C2Ab cluster includes Burkholderia sp. and Enterobacter sp. with medium/low P solubilization and PGPR traits. Cluster C2B was further divided into 2 subclusters: C2Ba and C2Bb, wherein C2Ba includes isolates showing medium P solubilization, medium/high phytohormone production, and medium/low siderophore and ammonia production. C2Bb further has two clusters, namely C2Bb1 and C2Bb2. The C2Bb1 cluster contains isolates with medium P solubilization and PGPR traits. These isolates were biochemically identified as Streptococcus sp., Bacillus sp., and Paenibacillus sp. Cluster C2Bb2 contains isolates biochemically identified as Bacillus sp., Paenibacillus sp., Arthrobacter sp., and Burkholderia sp., which exhibited high/medium P solubilization, medium/high phytohormone production, and medium/low siderophore and ammonia production. Thus, cluster C2Bb2 was the cluster with the most promising isolates in terms of PGPR traits.
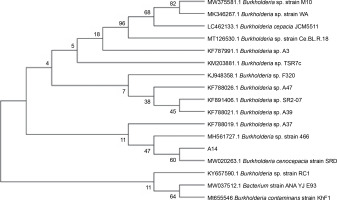
Among all the isolates of cluster C2Bb2, isolate A14 (Burkholderia) was the most promising isolate with high TCP solubilization, medium phytate mineralization, high enzyme production, medium/high phytohormone production, and medium ammonia production. This strain also showed nitrogen fixation activity, zinc solubilizing potential, and catalase production. This strain was identified as Burkholderia cenocepacia strain FDAARGOS_7 through the 16S ribosomal rRNA sequencing method. A similarity search analysis of 16S rRNA showed that the isolate was identical to “B. cenocepacia strain SRD” (99.71%). As shown in Figure 2, the neighbor-joining phylogenetic dendrogram based on 16S rRNA sequences and constructed using MEGA-X showed a relationship between the isolated strain and related B. cenocepacia sp. . The sequence obtained was deposited in the Gen-Bank nucleotide sequence database under the accession number MW775329. Different species of Burkholderia have been found to be associated with the plant rhizosphere. Several strains found in association with maize, sugarcane, tomato, and coffee are known to fix nitrogen (Paungfoo-Lonhienne et al., 2014). Burkholderia strains are considered to be agriculturally important because of their remarkable potential to mobilize phosphates and produce siderophores and phytohormones, thereby enhancing the plant’s resistance to biotic stress and acting as a biocontrol agent (Compant et al., 2008; Caballero-Mellado et al., 2007).
Conclusions
The study of the PGPR traits of the various isolates obtained in the present study showed that the B. cenocepacia strain possessed high TCP solubilization, medium phytate mineralization, high enzyme production, medium/high phytohormone production, and medium ammonia production. The results revealed that B. cenocepacia can serve as a potential candidate for the development of a biofertilizer. The study also concludes that PMB offer a better option to mobilize the trapped organic P reserves in soil. More studies should be conducted on the phytate mineralizing potential of rhizospheric bacteria from different rhizospheric soils. Future studies should focus on the development of B. cenocepacia biofertilizer and on the study of its effect on different crops in field conditions.