Introduction
Cyanobacteria are recognized as the earliest photosynthetic prokaryotes that existed on Earth. They commonly assimilate CO2 from atmospheric gases, which amounts to approximately 3 × 1014 g of C (carbon) (Garcia-Pichel et al., 2003) via), through photosynthetic machinery (Arrigo, 2005). The carbon concentrating mechanism (CCM) is responsible for fixing atmospheric carbon dioxide via carbonic anhydrase (CA), a zinc (Zn) metalloenzyme that is universally present among plants, microbes, and animals (Mondal et al., 2016; DiMario et al., 2017; Aspatwar et al., 2018). CA and inorganic carbon transporters are essential for the CCM in cyanobacteria, which increases the CO2 concentration in the vicinity of Rubisco (Price et al., 2008). Cyanobacteria containing defective CAs displayed abnormal growth (Price et al., 1989). CAs are encoded by three evolutionarily unrelated gene families, α CAs, β CAs, and γCAs. The α CAs are mainly found in vertebrates, plants, algae, and eubacteria; β CAs in bacteria, algae, and fungi; and γ CAs in archaea and bacteria. Additionally, marine diatoms have δ and ζ CAs, while protozoa express η CAs (Chegwidden et al., 2000; Supuran, 2010,2008; Frost and McKenna, 2013; Imtaiyaz et al., 2013). Zn is the metal ion present in the active site of almost all CAs, coordinated by three amino acid residues and one water molecule, which ionizes into an OH−ion. A hydrogen bond links this OH− ion to the amino acids present in the active centre. α-CAs and Γ CAs have HIS as the conserved amino acid, whereas β CAs have CYS2HIS(X) residues arranged in a tetrahedral formation, where X represents an Asp residue or any substitutable ligand (Iverson et al., 2000; Strop et al., 2001; Supuran, 2004; Sawaya et al., 2006). CA stimulates the fixation of CO2, which is carried out by a nucleophilic attack with the hydroxide ion bound to the Zn atom (Sayre, 2010).
The abundant growth of various algae and cyanobacteria in freshwater and marine water bodies frequently poses consequential threats as they are often associated with harmful toxins (Sivonen and Jones, 2009). Cyanobacterial blooms are known to contain hepatotoxins (microcystin andnodularin), cyclic heptapeptides, neurotoxins (anatoxin-a and homoanatoxin-a), and alkaloid cytotoxin (cylindrospermopsin). When the cyanobacteria mature, they lyse and release these toxins into their surroundings (Dittmann and Wiegand, 2006; Sivonen and Jones, 2009). Biofilm-producing cyanobacteria colonize various historical monuments, caves, natural/ artificial stones (lime mortar, carbonate, and sandstone), and pre-historic sites via an adhesion mechanism (Albertano, 2012; Popović et al., 2018). These biofilms are stress-tolerant and can adapt to high temperatures, desiccation, humidity, and various other factors. They also secrete biogenic pigments that cause staining and deterioration of stone surfaces (Albertano, 2012). Thus, microbial toxins significantly reduce the aesthetic value of prehistoric monuments, attributable to their acidolytic and oxido-reductive potential leading to erosion (Gaylarde, 2020). Although pesticides can inhibit cyanobacterial biomass (Nirmal Kumar et al., 2011; Debnath et al., 2012; Ni et al., 2014), continuous usage of these chemicals has harmful eco-toxicological impacts on the environment, along with various health issues and the death of non-targeted microorganisms (Singh et al., 2020). Herbicides such as diuron release toxic substances upon degradation, which are highly persistent and affect aquatic species (Field et al., 2003). Endothall is toxic to cyanobacteria, yet its application as an algaecide is not preferable due to its toxicity on zooplankton (Holdren, 2001; Osano et al., 2002; Giacomazzi and Cochet, 2004).
This study aimed to create a phytochemical database to discover lead molecules that could act as a possible inhibitor, targeting CA without adverse effects on nontargeted inhabitants. CA is the predominant enzyme that helps in increasing the biomass of cyanobacteria, as suggested by various studies (Jiang et al., 2014; Mondal et al., 2016; Tsuzuki et al., 2019). Molecular docking via in silico is a suitable approach to carry out this study (Gupta et al., 2013; Suganya et al., 2017). This method is a convenient tool to predict the binding affinity of the leads to the active site of the desired protein based on strong binding docking scores (Muhammad and Fatima, 2015; Kwatra et al., 2021). Furthermore, density functional theory (DFT) calculations provide higher occupied molecular orbital (HOMO), lower occupied molecular orbital (LUMO) and frontal orbital (FO), which give a preliminary idea about the reactivity and stability of the molecules (Parr et al., 1922). Additionally, the pk CSM web server was used to calculate the absorption, digestion, metabolism, excretion, and toxicity (ADMET) of the compounds, while SwissADME and Molsoft L.L.C online web servers were used to determine the pharmacokinetic properties of small molecules (Molsoft LLC., 2007; Pires et al., 2015).
Materials and methods
Preparation of phytochemical database
In this study, native medicinal plants belonging to Odisha (listed in Supplementary Table S1) were selected to identify lead molecules by targeting the CA (a metalloprotein enzyme) of cyanobacteria. Phytochemicals extracted from these medicinal plants (Supplementary Table S1) were searched in PubChem (https://pubchem.ncbi.nlm.nih.gov/), while those without any previous PubChem history were searched in the scientific literature (Venkatesh et al., 2014; Arunachalam et al., 2015; Panigrahi et al., 2015; Sajini et al., 2019; Jena et al., 2020). The two-dimensional structure of these phytochemicals was drawn using ChemDraw pro-8.0.1 and then converted into a three-dimensional (3D) structure using Chem3D Ultra 8.0. The structures were optimized by adding partial charges using the protonated 3D module, and their energies were minimized using the molecular operating environment (MOE) 09 software (El-Azab et al., 2010; El-Deeb, 2010). The NPACT (naturally occurring plant-based anticancer compound-activity-target) database, which consists of 1 800 molecules, was also incorporated with the phytochemicals present in Odisha. Finally, a phytochemical database containing approximately 3K molecules was created (Supplementary Table S1) by sorting them one by one in the molecular database file using MOE software.
In silico ADMET investigations
The energy-minimized molecules were converted into Simplified Molecular Input Line Entry System (SMILES) format and submitted to online web servers such as pk CSM (http://biosig.unimelb.edu.au/pkcsm/), Swiss-ADME (http://www.swissadme.ch/), and Molsoft L.L.C. (http://molsoft.com/) to predict the ADMET properties of the molecules present in the prepared database. These search engines predicted the pharmacokinetics of the molecules using the bioavailability radar, which considers six physicochemical properties, including lipophilicity, size, polarity, solubility, flexibility, and saturation, to detect toxicity (Molsoft LLC, 2007; Pires et al., 2015). The Lipinski filter, pioneer rule-of-five, was also used in this tool to predict drug likeness (Tripathi et al., 2019). Further studies were conducted on the molecules that fulfilled the above-mentioned criteria.
DFT optimizations
The lead candidates obtained from the ADMET filter were sketched using ChemDraw pro-8.0, and DFT optimizations were performed using the Becke 3-Parameter, Lee-Yang-Parr/Gaussian set (B3YLP/G*) level of theory. The HOMO and LUMO frontal orbitals were determined using the ORCA program 4.1.1. (Neese, 2012, 2017). The band gap in eV provided a preliminary idea about the nature of the molecule, such as its reactivity, hardness, softness, electronegativity, and global electrophilicity. These parameters were calculated using the equations given by Parr (1922).
Preparation of receptor protein
The 3D structure of beta-CA (Synechocystis sp. PCC 6803) was obtained from the online protein database (https://www.rcbs.org) with PDB-ID 5SWC, having a molecular weight of 162.76 kDa, an atom count of 11 602, and a resolution of 1.45 Å. The β-CA is a unique hexameric protein arranged in dimers of trimers in a three-fold symmetry axis. Refinement of this receptor protein was carried out by removing water molecules. The 3D protonation module was used to add nonpolar hydrogen atoms. Further, minimization was conducted by the MMFF94x force field, and the gradient was set at 0.05 using MOE 09 software. The binding site was predicted using ArgusLab 4.1 (Thompson, 2004).
Molecular docking simulation
To analyse the leading inhibitors of the CA enzyme, molecular docking was conducted using non-toxic leads from the 3K phytochemicals obtained from the PubChem database (Supplementary Table S1). The site finder module of MOE09 was used to identify the binding pocket, including the Zn301 atom at the base of the binding site. The docking scores of the ligands to the receptor were calculated using the following computations: rescoring 1: London-dG, placement: triangle matcher, retain: 10, refinement: forcefield, and rescoring 2: London-dG. The ligand molecules with the minimum binding scores were chosen based on their most relevant interaction with the target. The LigX module of MOE was used to identify the interactions with the receptor target.
Results
ADMET filter
The ADMET properties of all the phytochemicals were evaluated, and out of 3K molecules, 50 molecules passed all the criteria, having suitable ADMET profiles, accepting the Lipinski rule of 5, Veber rule, Muegge rule, Ghose rule, and Egan rule. Additionally, these phytochemicals did not exhibit herG-I/II cardiac muscle toxicities, AMES toxicity, blood-brain barrier, hepatotoxicity, and skin sensitization (refer to Fig. 1 and Table 1).
Fig. 1
ADMET analysis of a phytochemical database containing 3K molecules filtered using online search engines
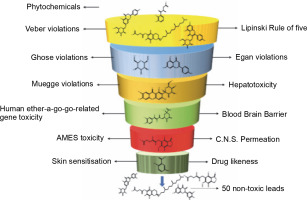
Table 1
ADMET properties of non-toxic molecules obtained from online search engines
Protein structure analysis of CA
The beta-CA of Synechocystis sp. PCC 6803 is arranged in dimeric form as A–B, C–D, and E–F chains (refer to Fig. 2). The alignment tool (BLOSUM40) of the MOE was used to determine the structural identity between the chains present in the X-crystallographic structure. Chain A was set as the reference template, and the remaining chains were superposed onto the reference template. All six chains of CA showed similarities in amino acid position, including loops, helices, alpha, and beta sheets. The superposed structures with the reference template (chain A) showed a pair wise root mean square deviations (RMSD) of each chain below 1Å (refer to Fig. 3). Finally, chains A–B and single chain A were selected for docking.
Fig. 2
The complete set of cyanobacterial carbonic anhydrases consists of six chains, with each chain represented by a distinct color: chain A (red), chain B (green), chain C (blue), chain D (violet), chain E (yellow), and chain F (cyan)
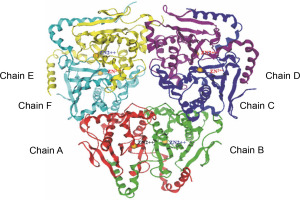
Fig. 3
(A) The reference template sequence was chain A, and the remaining chains B–F were superposed onto this reference; (B) RMSD was recorded
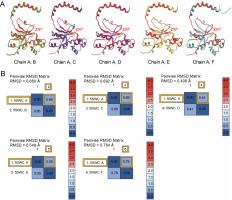
The RMSD of superposed chains B, C, D, E, and F onto the reference chain A were found to be 0.660, 0.692, 0.406, 0.549, and 0.784 Å, respectively.
Molecular docking analysis
Docking simulations of the retrieved 50 non-toxic phytochemicals (Table 1) were carried out at the active site containing the amino acids 103 ALA, 41 ASP, 39 CYS, 101 CYS, 84 ALA, 102 GLY, and 98 HIS along with Zn301 bound to CYS 101, CYS 39, and HIS 98 with the homolog chains A–B and single chain A. Based on the interaction with chain A and chains A–B, the non-toxic phytochemicals with top binding score (S score), drug-likeness score, and Zn interaction status were considered for this study (Table 2 and Table 4).
Table 2
Phytochemicals showing their binding (S-value) and drug-likeness scores with chain A
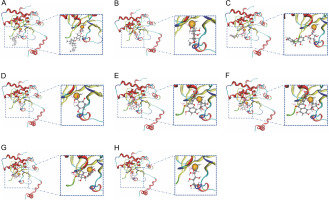
With single-chain A, most of the phytochemicals exhibited an interaction with Zn301 (CYS 101, CYS 39, and HIS 98). Ligands which demonstrated interaction with Zn301, also interacted with ASP 41 and other amino acids. Alpha-tocopherol succinate and 2,5-dihydroxybenzoic acid only interacted with ASP 41. Similarly, methyl alpha-D-glucopyranoside and methyl galactopyranoside demonstrated similar interaction with ASP 41, SER 42, and GLY 102 due to their structural similarity. The interaction of apigenin-7-glucuronide was observed with ASP 41, GLU 187, ARG 43, and GLY 102. Similarly, Orientin exhibited an interaction with ASP 41, GLY 102, LYS 105, and LYS 109, while dihydromyricetin interacted with ASP 41 and LYS 105. Galactitol showed interaction with ASP 41, SER 42, and GLY 102. Table 2 and Figure 4 depict the interactions, and Table 3 provides information on the distances of the Zn-interacted molecules.
Table 3
The interactions and distances displayed by leads with zinc and other amino acids with chain A
Fig. 4
The figure shows the molecular interaction between zinc ion in the binding pocket of beta-carbonic anhydrase chain A and the following compounds: (A) Alpha-tocopherol succinate, (B) Methyl alpha-D-glucopyranoside, (C) Apigenin-7-glucuronide, (D) 2,5-Dihydroxybenzoic acid, (E) Orientin, (F) Mycophenolic acid, (G) Dihydromyricetin, and (H) Galactitol
In the active site of chains, A–B, the ligands that demonstrated interaction with Zn301 were alpha-tocopherol succinate and mycophenolic acid through CYS 101, HIS 98, and CYS 39. Alpha-tocopherol succinate interacted with GLY102 of chain A and GLN 30 of chain B, while mycophenolic acid interacted with both ASP 41 and LYS 105 of chain A. Non zinc ligands interacted with amino acids in chain A (ASP A41, ARG A43, LYS A105, LYS A109, HIS A100, and GLY A102) and chain B (GLN B30, TYR B83, and ALA B87) as shown in Table 4 and Figure 5. Table 5 provides information on the distance score % of the Zn-interacted molecules.
Table 4
Phytochemicals showing their binding scores (S-value) and drug-likeness with chain A–B
Table 5
The interactions and distances displayed by leads with zinc and other amino acids with beta-carbonic anhydrase chain A–B
Fig. 5
The interaction shown by (A) Alpha-tocopherol succinate and (B) Mycophenolic acid with zinc ion in the binding pocket of beta-carbonic anhydrase chains A–B
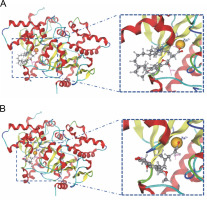
Molecular docking simulations revealed that alphatocopherol succinate and mycophenolic acid interacted with Zn301 and neighboring amino acids present in the binding site. These findings suggest that alpha-tocopherol succinate and mycophenolic acid could be effective phytochemicals that inhibit the cellular functioning of CAs found in cyanobacteria.
Global electrophilicity parameters of lead phytochemicals
The frontier molecular orbitals provide data on the chemical reactivity and physical parameters of molecules (Sheikhshoaie et al., 2014). The HOMO acts as an electron donor, and the LUMO acts as an electron acceptor. The energy gap between HOMO and LUMO determines the stability of structures, including their chemical reactivity and kinetic stability (Kim et al., 2013). The computed data on the HOMO, LUMO, energy gap (ΔE), ionization potential, electron affinity (A), chemical potential (μ), electron negativity (χ), hardness (η), softness (σ), and electrophilicity (ω) of the molecules are presented in Table 6 and Figure 6.
Table 6
Global electrophilicity parameters of alpha-tocopherol succinate, methyl alpha-D-glucopyranoside, apigenin-7-glucuronide, mycophenolic acid, 2,5-dihydroxybenzoic acid, orientin, dihydromyricetin, methyl galactopyranoside, and galactitol
Fig. 6
The energy shifting of HOMO and LUMO due to the occupancy of atoms present in the molecular scaffold of the following compounds: (A) Alpha-tocopherol succinate (B) Methyl alpha D-glucopyranoside (C) Apigenin-7-Glucuronide, (D) Mycophenolic acid (E) 2,5-Dihydroxybenzoic acid (F) Orientin (G) Dihydromyricetin, and (H) Galactitol
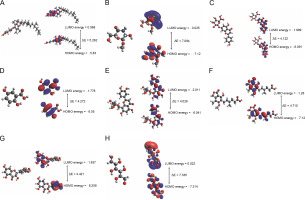
In alpha-tocopherol succinate, the energy shift was assigned to the succinate moiety, in LUMO to the tocopherol ring in HOMO. In apigenin-7-O-glucuronide, the energy shift was assigned to the apigenin moiety for both HOMO and LUMO. In mycophenolic acid, the energy shift was assigned to the phthalide intermediate in LUMO to 2-(but-2-enyl)-3-methoxy-methylphenol in HUMO. In 2,5-dihydroxy benzoic acid, the energy shift LUMO was assigned to the benzoic acid and HOMO to the hydroquinone moiety. In orientin, both LUMO and HOMO energy shift was assigned to the 2-(3,4-di-hydroxyphenyl)-5,7-dihydroxy-8-methyl-4H-chromen-4-one moiety. In dihydromyricetin, the energy shift in LUMO was assigned to the 3,5,7-trihydroxy-2,3-dihydrochrome-4-one moiety, and HOMO was assigned to benzene 1,2,3-triol. In galactitol, LUMO energy shift covered a vast surface of the galactitol molecule, whereas HOMO distribution was symmetrical. In methyl alpha-D-glucopyranoside and methyl galactopyranoside, LUMO was assigned to the methyl position, and HOMO was assigned to the overall molecule, as shown in Figure 6. Table 6 presents the band gaps of alpha-tocopherol succinate, apigenin-7-O-glucuronide, mycophenolic acid, 2-5-dihydroxybenzoic acid, orientin, dihydromyricetin, and galactitol as 5.262, 4.122,4.710, 4.272, 4.029, 4.421, and 7.335 eV, respectively, and the electrophilicity indices as 1.948, 3.940, 2.805,3.586, 4.022, 3.705, and 1.812 eV, respectively. Due to their structural similarity, methyl alpha-D-glucopyranoside and methyl galactopyranoside had an energy gap of 7.094 eV and an electrophilicity index of 1.798 eV. Overall, it was observed that all the molecules had high energy band gaps and a low global softness index. This suggests that the molecules are effective due to their stability and less reactivity.
Discussion
Cyanobacteria are primitive organisms that exist in various habitats, from hot springs to cold arctic regions. These bodies are gradually becoming eutrophied due to anthropogenic and industrial activities. Underdeveloped nations with limited technology sources rely directly on open water systems, lakes, ponds, rivers, oceans, and sanitary households for their day-to-day work. These water systems often contain cyanobacteria biofilms that secrete toxins, thereby deteriorating water quality on a large scale and leading to major health issues (O’Neil et al., 2012; Paerl et al., 2013). Therefore, measures should be taken to suppress harmful cyanobacterial growth. Naturally occurring nontoxic phytochemicals are generally secondary metabolites (terpenes, flavonoids, flavones, alkaloids, and polyphenols) that carry out several metabolic activities related to defense, stress tolerance, and interactions (Leitzmann, 2016). They are known as “nutraceuticals” due to their physiological benefits, such as being anticancer, antioxidants, anti-inflammatory, antiviral, and anti-bacterial agents (Brindha, 2016; Winter, 2017; Velmurugan et al., 2018; Ranjan et al., 2019). Natural compounds produced from plants, such as stilbenes, nostocarboline, berberine, and phenylpropanoid glucosides, have been reported as potent algaecides (Becher et al., 2005; Mizuno et al., 2008; Jančula et al., 2010; Zuo et al., 2018). Similarly, phytochemicals isolated from plants like Pistacia integerrima, brown alga Dictyopteris hoytii, flavonoids from Luffa acutangular (L.) Roxb, Murraya paniculata, and Padina pavonica,have potent CA inhibitory activity (Sarikaya et al., 2011; Sahin et al., 2012; Karioti et al., 2016; Chanda et al., 2019; Sangkaew et al., 2020; Irfan et al., 2021; Rafiq et al., 2021).
As the application of synthetic compounds to algal blooms can adversely affect aquatic organisms, we have chosen nontoxic phytochemical leads for this study that can inhibit blooms by targeting cyanobacterial beta-CAs without affecting non-targeted organisms.
A phytochemical database of 3K molecules was created, considering the medicinal plants of Odisha via a previously reported literature survey. The toxicity of phytochemicals in the database was investigated, and non-toxic phytochemicals were determined using multi software packages (Molsoft LLC., 2007; Pires et al., 2015). Fifty molecules (Table 1) from 3K molecules followed drug likenesses and ADMET properties (Pires et al., 2015; Tripathi et al., 2019). Among them, the two lead molecules, alpha-tocopherol succinate and mycophenolic acid, showed drug-likeness scores of 0.93 and 0.69, respectively, out of 1 (Molsoft LLC., 2007). The stability and reactivity of the lead molecules were determined by DFT calculations. The HOMO–LUMO energy gap was identified using B3YLP/G* level of theory, from which the ionization potential, electron affinity, chemical potential, electron negativity, hardness, softness, and electrophilicity were calculated. Hard molecules possess large energy gaps and are highly stable, whereas soft molecules have a smaller energy gap and are highly polarizable (Pilli et al., 2015). The energy gap, hardness, softness, and electrophilicity parameters of alpha-tocopherol succinate were 5.262, 2.631, 0.380, and 1.948 eV, respectively, and for mycophenolic acid, they were 4.710, 2.355, 0.424, and 2.805 eV, respectively, which supports the findings by Pilli et al. (2015).
These findings support a previous study demonstrating that phytochemicals play a pivotal role in inhibiting the biomass of cyanobacterial species, as demonstrated by both in silico and in vitro studies (Mir and Nayak, 2022).
The beta-CA of cyanobacteria Synechocystis sp. PCC 6803, with the cofactor Zn (Zn301) present in the active site, was chosen as the target protein. Zn functions as a catalyst and regulates enzymatic reactions (Escudero-Almanza et al., 2012). Initially, single chain A and later double chains A–B of beta-CA were used for docking to identify interactions with Zn2+. Alpha-tocopherol succinate and mycophenolic acid were found to be potent phytochemical leads as they exhibited interactions with Zn2+ and its coordinate amino acids CYS 101, CYS 39, and HIS 98 in both single and double chain. Previous reports have shown that phytochemicals such as pistacide-A and pistacide-B obtained from P. integerrima and lacceroic acid from brown alga D. hoytii is potent inhibitors of CA-II (Irfan et al., 2021; Rafiq et al., 2021). These inhibitors showed binding scores of −3.45, 3.64, and −9.53 Kcal/mol, respectively, against the reference molecule acetazolamide (−9.47 Kcal/mol) (Irfan et al., 2021; Rafiq et al., 2021). Similarly, triazine substituted sulfamerazine-derived compound 5 (4-ethoxy) and compound 8 (4-bromo) have also been identified as potent human CA I and II inhibitors, respectively (Bilginer et al., 2021). Compound 5 had a binding score of −7.36 Kcal/mol with human CAI (hCAI) and compound 8 (−8.26 Kcal/mol) with hCAII (Bilginer et al., 2021), respectively.
In this study, the identified most potent phytochemicals, alpha-tocopherol succinate, and mycophenolic acid, possessed binding scores of −9.23 and −14.41 Kcal/mol, respectively, which were lower than those of the reported CA II inhibitors (pistacide-A, pistacide-B, and lacceroic acid) and hCAI/hCAII inhibitors (compounds 5 and 8). This confirms the strong interaction of the lead phytochemicals with the active site of beta-CAs.
Several phytochemicals have been reported to act as anti-carbonic anhydrase agents against CA-II, CA-V, hCA-IX, and hCA-XII, which are associated with abnormalities such as tumor progression, cancer, obesity, and Alzheimer’s disease (Rahman et al., 2015; Queen et al., 2018; Fois et al., 2020; Mahmud et al., 2021). However, to our knowledge, no studies have been conducted targeting cyanobacterial beta-CA. Therefore, in this study, we have considered phytochemicals that showed robust bindings with the amino acids and co-factor present in the binding site of beta-CAs. These phytochemicals have the potential to inhibit the cellular functioning of CA, thus restricting the allocation of bicarbonates to the RuBisCo site and curtailing the formation of cyanobacterial biomass. Further studies such as molecular dynamics simulations and in-vitro experiments will be carried out to validate the present in-silico findings.
Conclusion
In rural areas and economically disadvantaged countries, people rely on open-water systems for their daily needs. Unfortunately, anthropogenic activities, such as the disposal of leached micronutrients and industrial effluents into aquatic environments, lead to cyanobacterial blooms. These harmful cyanobacteria release cyanotoxins into the water, which can cause major health issues like hepatotoxicity and immunotoxicity when ingested orally. As such, it is important to identify novel anti-cyanobacterial agents that do not have any negative side effects on aquatic organisms. In this study, alphatocopherol succinate and mycophenolic acid phytochemicals were found to exhibit binding interactions with the co-factor Zn in both single chain A and complexed chains A–B. These lead molecules interact with the Zn-binding site present in the enzyme and may inhibit the functioning of the beta-CA complex, thereby restricting biomass generation. Considering the present and future perspectives of water scarcity, this approach may be a possible way to protect the biotic environment, water bodies, and ancestral monuments. Further in vitro experiments on lead phytochemicals are necessary to validate the present in silico study.










