Introduction
Colorectal cancer (CRC) is the fourth leading cause of death due to cancer in the US [1]. Worldwide, there is an approximately 4.3% risk of CRC in men and women [2]. The death rate due to colon cancers has increased [3]. In the early 1990s, the death rate was 2.87 per 100,000, and it nearly doubled to 6.8 in 2017 [4]. The annual report of the National Cancer Registry of Iran (INCR: National cancer Registry in Iran) indicates CRC as the fourth most common cancer in males after stomach, bladder, and prostate [5]. The central, western and northern provinces of Iran have the highest incidence of CRC, while the southeastern provinces have the lowest incidence [6]. Several factors such as age, family history, and diet affect the occurrence of CRC [7].
According to studies, nearly 250 different mutations have been identified in patients with CRC, which is approximately equivalent to 55% of the known mutations related to mismatch repair (MMR) genes [8]. Abnormal methylation as a regulatory mechanism of gene expressions can be seen in tumour suppressor genes in various cancer types, including such as in [9]. Different epigenetic biomarkers are used for the diagnosis and prediction of prognosis of CRC [10]. Methylated septin 9 (mSEPT9) showed extremely high levels of methylation in CRC tissues, is used for CRC serological diagnosis, and has increased sensitivity [11].
CRC has rising incidence and mortality; thus, accurate and early diagnosis is important [12]. Colonoscopy is now the most direct technique to detect colorectal neoplasm [13]. Nonetheless, colonoscopy is not acceptable because of its possible risk and discomfort [14]. Non-invasive blood testing is easy to use with the possibility of frequent monitoring following diagnosis and treatment [15]. In a study in Germany, 63.4% (109/172) of cases did not accept screening colonoscopy, while 82.6% (90/109) selected blood testing [16]. The US Food and Drug Administration (FDA) recently approved detection of circulating mSEPT9 DNA in blood [17], which provided a commercially available alternative non-invasive test for the CRC screening [12]. Carcinoembryonic antigen (CEA), an oncofoetal antigen, is produced epithelial tumours of endodermal origin [18]. According to the American Society of Clinical Oncology, it should be assessed every 3 months for a minimum of 3 years in CRC patients in stages II and III after surgery [19]. Many studies have evaluated the diagnostic value of peripheral blood mSEPT9 to detect CRC; however, the diagnostic accuracy of CRC is highly varied [17, 20]. As a result, this study was conducted with the aim of investigating CEA and septin 9 in patients with CRC.
Methods
Aims
We evaluated the specificity and sensitivity of septin 9 and carcinoembryonic antigen in patients with CRC.
Design
The Cochrane instructions [21], the PRISMA statement [22], and synthesis without meta-analysis (SWiM) guidelines were used for a systematic review of empirical quantitative studies [23].
Search methods
Inclusion criteria
The MEDLINE (PubMed interface), Psych-Info, Cochrane, and CINAHL (EBSCO interface) databases were searched between January 2012 and December 2022. MESH words were added to free text terms using a coherent search strategy: marker (CEA, septin 9, surgical, sensitivity, specificity) and cancer (CRC).
We developed the search strategy for Medline followed by the other databases. More studies were found by searching the references of the selected studies, and it was the final stage to add the articles.
Primary human studies in the period 2012-2022 that had CRC, studies that evaluated the sensitivity and specificity of CEA and septin 9 serum markers in CRC, published in English during 10 or feweer years ago, and assessing patients 18 years or older were considered.
Study selection
After choosing 2 researchers based on recent studies from databases from January 2012 to December 2022, the studies were chosen according to the first 100 abstracts obtained from the databases. Then, 2 different reviewers evaluated other abstracts, and in cases of uncertainty regarding including the abstracts, the full texts were read and relevant abstracts published from January 2012 were included considering the inclusion criteria. Some full-text articles not accepted by the first reviewer were reassessed by the second reviewer considering the exclusion criteria. Two independent reviewers assessed the eligibility. Also, case-control studies were not included.
Search outcome
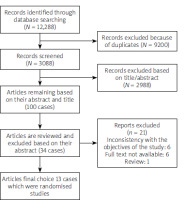
A total of 12,288 records were detected. Duplicates (N = 9200) were not included. Based on the inclusion criteria, 3088 abstracts were reviewed, and extraction and review of 2988 abstracts were done. Also, 100 full-text articles were detected based on their abstract and title between January 2012 and December 2022. Also, 34 related published texts were found, but after reading the full texts, 21 articles were excluded because of reports excluded, inconsistency with the objectives of the study (n = 6), full text not available (n = 6), and being a review article (n = 1). Finally, 13 articles remained (Figure 1).
Quality evaluation and data extraction
The Quality Assessment Tool [24] assessed the quality of the quantitative articles.
The quality of 34 articles was assessed using the STROBE list (Table I), and 13 appropriate articles were included. The full-text studies were evaluated prior to data extraction. This tool is adjusted based on a tool designed by the Effective Public Health Practice Project [25] applied in many systematic reviews [26, 27].
Table I
STROBE Statement – checklist of items that should be included in reports of observational studies
The information were extracted as follows: author; country; year; age; number of patients; sex; cancer type; study type; sensitivity and specificity of septin 9; sensitivity and specificity of C; CEA measurement method; septin 9 (Table II) [28–39]. Two authors extracted data from each article, and in the case of disagreement a third opinion was used.
Table II
Characteristics of the studies
| No. | Author | Year | Country | Patients with CRC | Sex | Ageaverage | Cut off Of CEA | CEA (positive) % | Septin 9 (positive) % | Type of manuscript | Correlation between tumour grade and CEA.Septin9 levels | Time | Sample | Measurement of Septin9 | Measurement of CEA | Septin9 | CEA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| After surgery | Before surgery | Sen % | Spe % | % Add | NPV % | AUC % | Accuracy % | Sen % | Spe % | % Add | NPV % | AUC % | Accuracy % | ||||||||||||||||
| Male | Female | ||||||||||||||||||||||||||||
| 1 | Ma [47] | 2019 | China | 117 | 68 | 49 | 67.3 | > 3 | 48.2 | 73.2 | Prospective study | Increases | * | Plasma | PCR | Enzyme-linked ionosphere | 71.8 | 66.7 | 71 | 50.5 | 78.9 | 60.5 | |||||||
| 2 | Toth [29] | 2012 | Germany | 187 | - | - | 67.8 | > 4.3 | 14.2 | 88.9 | Prospective study | In stage 3 is 35.35 | * | Plasma | PCR | Cobas, Roche Diagnostics | 95.6 | 84.8 | 86.3 | 95.1 | 90.2 | 51.8 | 85.2 | 77.8 | 63.9 | 68.5 | |||
| 3 | Lu [39] | 2022 | China | 326 | 211 | 115 | 58.6 | > 3 | 66.7 | 73 | Case control study | In stage 4 is 81.8 | * | Blood | PCR,Epi proColon | Cobas, Roche Diagnostics | 77 | 88 | 88 | 75 | 82 | 82 | 73 | ||||||
| 4 | Leung [40] | 2019 | Hung Kong | 187 | - | - | 66.1 | > 3 | 48.2 | 73.9 | Prospective study | Stage 4 is 100% | * | Plasma | PCR,Epi proColon | Enzyme-linked ionosphere | 73.5 | 72.5 | 81.9 | 62.2 | 73.3 | 47.9 | 79.3 | 80 | 47.9 | 60 | |||
| 5 | Lee [41] | 2013 | South Korea | 197 | - | - | 65.3 | > 3 | 44.7 | 64.7 | Case control study | Stage 4 is 64.7% | - | * | Plasma | Realtime, PCR | Roche Diagnostic Corp | 36.6 | 90.6 | 80.4 | 57.6 | 62.9 | 20.8 | ||||||
| 6 | Eldeeb [42] | 2020 | China | 90 | 55 | 35 | 53.6 | > 2.9 | 15.8 | 38.4 | Prospective study | - | * | Blood | Sunred ELISA | Electrochemiluminescence immunoassay | 84.9 | 78.9 | 75 | 87 | 91.1 | 81 | 78 | 76 | 76 | 77 | 80.8 | 77 | |
| 7 | Sun [43] | 2019 | China | 557 | - | - | 65.2 | > 3.2 | 61.9 | 71.4 | Comparative study | - | * | Plasma | Epi proColon, PCR | Electrochemiluminescence | 73 | 94.5 | 63 | 96.5 | 83.5 | 52.4 | 91.8 | 65.4 | |||||
| 8 | Song [44] | 2019 | China | 120 | 72 | 48 | 61.4 | > 3 | 44.2 | 86.7 | Comparative study | - | * | Plasma | Epi proColon, PCR | Electrochemiluminescence | 86.7 | - | 44.2 | - | |||||||||
| 9 | Arellano [12] | 2022 | Spain | 32 | 16 | 12 | 50.75 | - | - | 53.5 | Prospective study | Increases | * | Plasma | Epi proCo-Enzyme-linked Ion, PCR ionosphere | 55.6 | 100 | 100 | 23.5 | 59.3 | |||||||||
| 10 | FU [33] | 2018 | China | 351 | 200 | 151 | 60 | > 3 | 50 | 61.2 | Prospective study | Stage 4 is 87.5 % | * | Plasma | Epi proColon, PCR | Electrochemiluminescence immunoassay | 61.2 | 98.4 | 93.7 | 86.8 | 80.2 | 88 | |||||||
| 11 | Wu [45] | 2016 | China | 291 | 183 | 108 | - | > 3 | 41.3 | 77 | Prospective study | - | * | Plasma | Epi proColon, PCR | Electrochemiluminescence | 76.6 | 95.9 | 94.9 | 80.6 | 88.2 | 86.3 | 41.3 | ||||||
| 12 | Yuan [46] | 2016 | China | 187 | 100 | 87 | 63.8 | > 0.7 | 32.9 | 74.9 | Prospective study | - | * | Plasma, tissue | PCR | Electrochemiluminescence | 62.6 | 91.7 | 92.8 | 58.8 | 77.7 | 73.3 | 32.9 | ||||||
| 13 | Ma [37] | 2021 | China | 103 | - | - | 67.3 | > 3 | 69.4 | 67.9 | Prospective study | stage 3 is 76.7 % | * | Plasma, tissue | PCR | Enzyme-linked ionosphere | 73.7 | 50 | 81.8 | 38.1 | 71 | 67.7 | 81.2 | ||||||
Statistical analysis
Homogeneity of findings was assessed through the results of inconsistency index (I2) value and Cochran’s Q test. For each study, false positive (FP), true positive (TP), false negative (FN), and true negative (TN) values were calculated according to the study data. A random-effects model calculated the overall effect. Forest plots containing descriptions of the results were applied to explain the estimates of the accuracy measures (specificities, sensitivities, negative and positive likelihood ratios (LRs), and diagnostic odds ratios (dOR), receiver operating characteristics curve (ROC) to describe the relationship between sensitivity and specificity of the test) with 95% confidence intervals (CIs). An area under the curve (AUC) close to one indicates good diagnostic performance of the test. Meta-Disc 1.4 was employed for all statistical analyses.
Results
Overall, 13 quantitative studies were analysed. Table II indicates the studies’ characteristics, which were all cross-sectional, conducted in China (N = 9), Spain (N = 1), Germany (N = 1), Hung Kong (N = 1), and South Korea (N = 1). Nine of these studies were prospective study studies, 2 were case-control studies, and 2 were comparative studies. The tested sample was reported in 9 studies on plasma, 2 studies on whole blood, and 2 studies on plasma and tissue. The total number of participants with colon cancer was 2745, and the patients’ mean age was 62.26 years. The mean level of CEA positivity was 44.79% and septin 9 was positive in 69.59%. In 8 studies, there was an incremental relationship between tumour grade and CEA, and septin 9 level was observed in cancer level 3 and 4. In 6 studies, sampling was done before colectomy surgery, and in 7 studies after it. In most studies, the septin 9 measurement method was by Epi proColon.
Overall accuracy measurements for septin 9 as a predictor for colorectal cancer detection
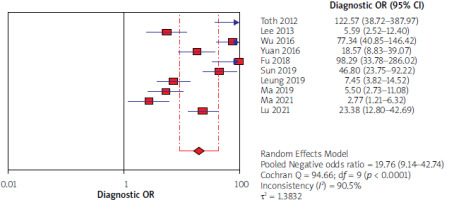
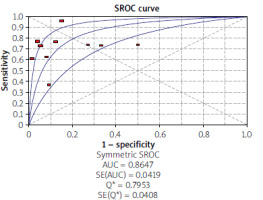
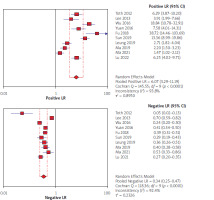
Using the results of 10 articles for septin 9, the total specificity and sensitivity were, respectively, 91% (95% CI: 90–92) and 71% (95% CI: 68–73) (Figure 2). The pooled positive and negative likelihood ratios (LRs) were, respectively, 6.07 (95% CI: 3.29–11.19) and 0.34 (95% CI: 0.25–0.47) (Figure 3). The pooled diagnostic odds ratio (dOR) based on the studies was high, at 19.76 (95% CI: 19.14–42.74) (Figure 4). The summary receiver operating characteristic (sROC) for septin 9 was 0.84 (Figure 5).
Figure 2
Forrest plot of sensitivity and specificity from accuracy studies of septin 9 for detection of colorectal cancer
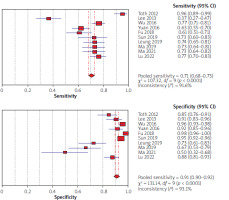
Figure 3
Forrest plot of likelihood ratios for positive and negative test results from studies of septin 9 for detection of colorectal cancer
Overall accuracy measurements for carcinoembryonic antigen (CEA) as a predictor for colorectal cancer detection
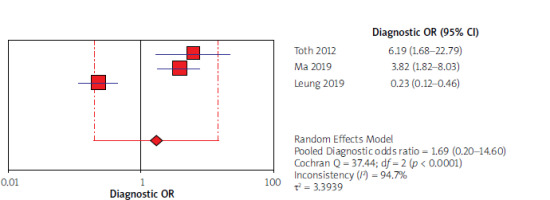
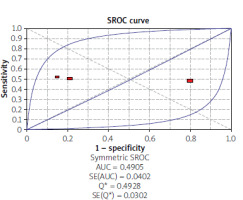
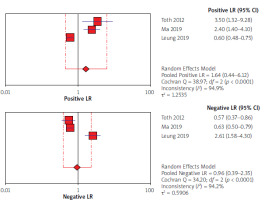
Using the results of 3 articles for CEA, the overall specificity and sensitivity were, respectively, 53% (95% CI: 45–61) and 49% (95% CI: 43–56) (Figure 6). The pooled positive and negative likelihood ratios (LRs) were also, respectively, 1.64 (95% CI: 0.44–6.12) and 0.96 (95% CI: 0.39–2.35) (Figure 7). The pooled diagnostic odds ratios (dOR) based on the studies were high, at 1.69 (95% CI: 0.20–14.60) (Figure 8). The summary ROC (sROC) for CEA was 0.49 (Figure 9).
Figure 6
Forrest plot of sensitivity and specificity from accuracy studies of carcinoembryonic antigen (CEA) for detection of colorectal cancer
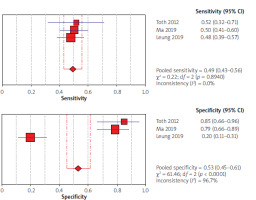
Figure 7
Forrest plot of likelihood ratios for positive and negative test results from studies of carcinoembryonic antigen (CEA) for detection of colorectal cancer
Discussion
CRC is the third most common cancer in women and men [40]. According to estimates, about 1.2 million new CRC patients are diagnosed, and nearly 608,000 deaths due to CRC are reported every year [41]. Despite advancements in CRC treatment, the CRC advanced stage at diagnosis can lead to a very undesirable prognosis [42, 43]. Patient prognosis is improved by using screening tests, and these tests predict long-term survival by detecting tumours at early stages, resulting in reduced mortalities related to CRC [44]. Assessment of tumour markers in serum, like CEA, has a limited specificity and sensitivity for CRC diagnosis [45]. The septin 9 test can assess the SEPT9 gene methylation status, which is hypermethylated in CRC patients [37]. Recently, the SEPT9 gene methylation assay, as a blood-based test, was used for detection and screening of CRC [46, 47]. The hypermethylated SEPT9 gene is a biomarker to diagnose CRC in tumoral tissue and peripheral blood [12].
This systematic review attempts to assess carcinoembryonic antigen and methylated septin 9 for serological diagnosis and monitoring of CRC patients. Among the various methylated genes, septin 9 is used for the serological diagnosis of CRC, characterised by high sensitivity, and CEA measurement has also been recommended in cancer patients after surgery [36, 48, 49]. We included 13 studies. We obtained more definite results by the descriptive combination of studies.
Assessment of biomarkers for the CRC diagnosis has a significant effect on its prognosis [50]. In general, in the review of the articles used in the current research, in 10 articles, the specificity and sensitivity of septin 9 were reported as 71% and 91%, respectively, and using the findings of 3 articles, the sensitivity and specificity of CEA were reported as 49 and 53%, respectively. This seems to indicate the importance of septin 9 as a biomarker in the diagnosis of CRC. Ma et al.’s study, using mSEPT9 methylation status to diagnose CRC, showed a significantly higher sensitivity than using CEA levels (73.2 vs. 48.2), and in general, in this study, mSEPT 9 was more sensitive for CRC diagnosis than was CEA and the combination of CEA, and mSEPT9 was more accurate [39]. Toth et al. declared similar findings, with respective specificities of 84.8% and 85.2% and sensitivities of 95.6% (88/92) and 51.8% (14/27) for mSEPT9 and CEA [29]. Also, mSEPT9 showed higher diagnostic value compared to CEA for sensitivity (73% vs. 52.4%) and specificity (94.5% vs. 91.8%) [34].
In the review of studies, we concluded that the sensitivity of the 2 tests increases with higher tumour staging. In the study by Ma et al., sensitivity showed an increase with increasing tumour staging (p = 0.04 and 0.04, respectively) [28]. In the study of Fu et al., the highest sensitivity was reported in stage 4 (87.5) [36]. In Lu et al.’s study, the sensitivity and specificity of both CEA and mSEPT9 tests increased in stages 3 and 4 more than stages 1 and 2 [30].
An AUC of close to one indicates a good diagnostic performance of the test. In our review, the AUC was reported as 0.84 for septin 9 and 0.49 for CEA. In the study of Eldeeb et al., the area under the curve was reported as 0.911 for septin 9 and 0.808 for CEA [33]. In Ma’s study, The accuracy regarding AUC of the methylated ratio and abundance for diagnosing CRC was nearly 0.71, which was more important than the unfavourable single methylated concentration performance (AUC = 0.55). The most favourable sensitivity for the detection of CRC was nearly 74% with a specificity of 50% for methylated abundance. The combined CEA and SEPT9 methylated abundance improved the performance with AUC to 0.92 [39].
In our study forest plots could explain the estimates of the accuracy measures – sensitivities, specificities, negative and positive likelihood ratios (LRs), and diagnostic odds ratios (dOR). The index of pooled positive and negative likelihood (LRs) for septin 9 was 6.07 against 1.64 for CEA and 0.34 for septin 9 against 0.96 for CEA. The index of pooled diagnostic odds ratios (dOR) was 19.76 for septin 9 and 1.69 for CEA. All these results obtained from the indicators indicate that septin 9 has a better and stronger diagnostic performance in identifying CRC. In the study of Xie et al. from 2019, according to the sensitivity and specificity indices and AUC, septin 9 biomarker was introduced as the best and most sensitive in detecting CRC [51]. The same results were found in the studies of Lu [30] and Allerano [12].
In most of the conducted studies, septin 9 measurement method was Epi proColon. The initial versions of the Epi proColon test (Epi proColon 2.0) possibly increased the SEPT9 performance in the diagnosis of CRC. The Epi proColon test, as a novel CRC screening test based on blood, can recognise the mSEPT9 (septin 9) gene in cell-free DNA collected from plasma [52]. In the study of Fu and Arellano, the (Epi proColon 2.0) method was also used [12].
Conclusions
In this review study, we investigated the expression level of septin 9 and carcinoembryonic antigen to identify CRC. We found that there is a direct relationship between the expression level of these genes and the grading of the cancer stage, so that in some studies, stages 3 and 4 showed the highest levels of gene expression. Strong evidence showed that septin 9 can be used as an effective marker for the detection and prognosis of CRC, and it also showed better differentiation than CEA.