Introduction
Animal sources of proteins, which are used to complement starchy diets predominantly consumed in developing countries, have become unaffordable to low-income families, and this has necessitated the search for cheaper sources of proteins (Kolapo and Oladimeji, 2008). Previous studies have reported the use of legumes as sources of milk and milk products to provide milk-like products in communities where milk is in short supply to probably help in reducing the incidence of protein deficiency diseases such as Kwashiorkor and malnutrition (Gesinde et al., 2008).
One such legume is soybean, a wonderful nutritional gift of nature. Soymilk is an off-white, aqueous, creamy liquid extract of soybeans, which resembles cow milk in both appearance and consistency. It has a high nutritive value and contains proteins, fats, carbohydrates, vitamins, and minerals (Adebayo-Tayo et al., 2008). Soymilk is popular because it plays an important role in the dietary pattern of people in most developing countries, coupled with its comparative low cost of production and high nutritional value. The nutrient content of 8 ounces of plain soymilk is 140 g calories, 10 g protein, 4 g fat, 14 g carbohydrate, 120 mg sodium, 1.8 mg iron, 0.1 mg riboflavin, and 80 mg calcium (Cruz et al., 2007). It has approximately the same amount of protein as that in cow’s milk, with a slight difference in the amino acid profile (Hajirostamloo, 2009; Mazumder and Begum, 2016). The health benefits of soymilk are well documented and include the presence of low cholesterol and lactose levels, making it easily consumable by individuals suffering from lactose intolerance; capable of reducing bone loss and alleviating menopausal symptoms, and preventing and reducing heart diseases and certain cancers (Stanley et al., 2014).
Soymilk is produced by adopting a traditional Chinese method with some modifications. The soybeans are sorted to remove stones and damaged or deformed seeds and washed with clean water. Whole soybeans are soaked in water overnight and ground with fresh water (water : bean, 8 : 10), and the slurry obtained is filtered through a muslin cloth. The residue, known as soy pulp or okara, is separated, and the filtrate is boiled thoroughly before serving or bottling. It is then stored at an ambient or refrigeration temperature (Kohll et al., 2017). The modifications made to the traditional Chinese method were aimed to eliminate the bean-like flavor associated with soybean products through heat inactivation of lipoxygenase (LOX). This process increases its acceptability by consumers, especially the Western ones (Baysal and Demirdoven, 2007).
One of the most important groups of bacteria known to humans is the Enterobacteriaceae family that comprises 53 genera taxonomically (and over 170 named species), of which 26 genera are known to be associated with infections in humans. They are gram-negative, non-spore-forming bacteria and include important foodborne pathogens such as Arsenophonus, Biostraticola, Brenneria, Buchnera, Budvicia, Buttiauxella, Calymmatobacterium, Cedecea, Citrobacter, Cosenzaea, Cronobacter, Dickeya, Edwardsiella, Enterobacter, Erwinia, Gibbsiella, Hafnia, Klebsiella, Kluyvera, Leclercia, Leminorella, Levinea, Lonsdalea, Mangrovibacter, Moellerella, Morganella, Obesumbacterium, Pantoea, Pectobacterium, Phaseolibacter, Photorhabdus, Plesiomonas, Pragia, Proteus, Providencia, Rahnella, Raoultella, Saccharobacter, Samsonia, Serratia, Shigella, Shimwellia, Sodalis, Tatumella, Thorsellia, Trabulsiella, Wigglesworthia, Xenorhabdus, Yersinia, Yokenella, Salmonella, Yersinia enterocolitica, pathogenic Escherichia coli (including E. coli O157 : H7), Shigella spp., and Citrobacter (Farmer et al., 2007). Some members of this family are regarded as opportunistic pathogens, such as Klebsiella spp., Serratia spp., and Citrobacter spp., which are implicated in the spoilage of fruits, vegetables, meats, poultry, eggs, milk, dairy products, and fish, contributing significantly to economic losses of valuable raw materials in agro-allied and food industries (Baylis et al., 2011).
Antibiotic resistance is described as a phenomenon that occurs when a microorganism that was previously susceptible to an antibiotic agent has now become resistant due to intrinsic features of its physiology or biochemistry (Levy and Marshall, 2004; Dugassa and Shukuri, 2017). Although antimicrobial resistance is not new, recently, there has been an increase in the number of resistant bacteria worldwide, together with reports of a wide range of resistance in single microorganisms (Levy and Marshall, 2004; Skrahina et al., 2013; Sabtu et al., 2015). Iwu et al. (1999), Roberts et al. (2009), and Blair et al. (2015) reported that epidemics due to drug-resistant bacteria are a serious global concern related to public health. The global emergence of multidrug-resistant (MDR) bacteria is a serious threat to the treatment of bacterial infections as it limits the effectiveness of antibiotics (Hancock, 2005). Most of these multidrug gram-negative and gram-positive bacteria are highly pathogenic to humans, and their ability to demonstrate this phenomenon is dependent on factors such as formation of biofilms and presence of efflux pumps and multidrug resistance proteins that contribute significantly to intrinsic and acquired resistance in these bacteria (Oluwatuyi et al., 2004).
According to Hawkey (2008) and Blair et al. (2015), antimicrobial resistance is currently the greatest challenge in the effective treatment of bacterial infections globally. For example, more than 70% of bacterial species implicated in hospital-acquired infections are resistant to at least one of the therapeutic antibiotics used to treat them; furthermore, more than 80% of food poisoning-causing bacteria such as Salmonella are reported to exhibit resistance to at least one type of antibiotics, and more than 50% of bacteria are resistant to two or more antibiotics (Hitoshi, 2006; Afolami and Onifade, 2018).
Food has been found to act as a carrier for the dissemination of antibiotic-resistant bacteria among humans, and the resistance exhibited by bacterial species belonging to Enterobacteriaceae complicates the treatment of many infections by producing plasmid-mediated enzymes known as extended-spectrum β-lactamases (ESβLs). Lavilla et al. (2008) reported that the production of these enzymes inhibits the action of some antibiotics such as cephalosporins, monobactams, and penicillins.
Plasmids are self-replicating extrachromosomal DNA molecules found in gram-negative and gram-positive bacteria. Morphologically, they are covalently bonded closed circular double-stranded DNA molecules; however, recent studies have reported the isolation of linear plasmids from different bacteria (Dib et al., 2015). Plasmids are structures that encode a variety of genetic determinants which enable bacteria to survive better in an adverse environment or to compete better with other microorganisms (Actis et al., 1999; Rozwandowicz et al., 2018). In addition, they are responsible for the dissemination of genes linked with antibiotic resistance. Svara and Rankin (2011) reported that when the resistance genes are mobile and carried on plasmids, the spread of antibiotic resistance is accelerated.
Soymilk is a popular milk product consumed widely by all age groups in the Ibadan Polytechnic Community, Nigeria, but it is mainly produced by local inhabitants. Recently, among the members of the Ibadan Polytechnic Community, incidences of protracted gastroenteritis were reported by consumers of hawked soymilk; this has necessitated the investigation of the occurrence and plasmid profiles of MDR Enterobacteriaceae isolated from hawked soymilk samples collected from the Ibadan Polytechnic Community, Oyo State, Nigeria.
Materials and methods
Sample collection
Ten soymilk samples were purchased from hawkers operating in the Ibadan Polytechnic Community (Polytechnic North, Polytechnic South, Polytechnic East, Polytechnic West, and Polytechnic Central). The soymilk samples were transported under hygienic conditions to the Food Biotechnology Postgraduate Laboratory, University of Ibadan, and stored at 4°C.
Isolation of bacteria from hawked soymilk samples
Ten milliliters of a soymilk sample was added to 90 ml sterile distilled water and homogenized. The samples were serially diluted following the method described by Fadahunsi and Makinde (2018) until a dilution factor of 10–6 was attained under aseptic conditions.
One milliliter of the diluted soymilk sample (10–2 dilution) was transferred to three different Petri dishes by using a sterile pipette. Twenty milliliters of plate count agar, MacConkey agar, and Salmonella-Shigella agar was added individually to the three Petri dishes. The procedure was repeated for 10–4 and 10–6 dilutions, and the plates were incubated aerobically at 37°C for 24 h. The plates were then examined for bacterial growth, and distinct representative colonies were subcultured repeatedly to obtain pure cultures, which were aseptically stored on agar slants in McCartney bottles and kept at 4°C for subsequent use.
Characterization of isolates obtained from soymilk samples
The bacterial species isolated from the samples were characterized macroscopically, microscopically, and biochemically by referring to Bergey’s Manual of Systematic Bacteriology (Sneath et al., 1986).
Macroscopic identification
The macroscopic or colony morphological examination was performed on the basis of physical characteristics of the isolates, such as cell shape, size, elevation, texture, color, edge, colony surface, and cell arrangement (Sneath et al., 1986).
Microscopic examination
Microscopic examination was performed to determine the cell shape of the isolated bacteria and their gram-staining characteristics.
Biochemical characterization of isolates
Biochemical tests such as catalase, oxidase, citrate, urease, and indole production; coagulase; methyl red; Voges-Proskauer test; sugar fermentation; motility test; spore staining; and hydrogen sulfide production were performed to determine the biochemical characteristics of the bacterial isolates (Kumar et al., 2012).
Antibiotic susceptibility of the bacterial isolates
The following antibiotics were used to determine the susceptibility pattern of the isolates: ertapenem (10 μg), gentamicin (10 μg), aztreonam (30 μg), amoxicillin-clavulanate (20/10 μg), ciprofloxacin (5 μg), ceftazidime (30 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), and cefpodoxime (10 μg) (Oxoid, England).
Preparation of McFarland Standard
The inoculum density was standardized for the susceptibility test by preparing a barium sulfate (BaSO4) solution of turbidity standard equivalent to 0.5 McFarland standard or its optical density equivalent and using it to compare with the turbidity of the inoculum (bacterial suspension).
The turbidity of the actively growing broth culture was adjusted with sterile saline to obtain a turbidity that was optically comparable to that of the 0.5 McFarland standard.
Antibiotic susceptibility testing procedure
Antibiotic susceptibility was tested using the disk diffusion method. A sterile cotton swab was dipped into the cell suspension in nutrient broth and seeded onto Mueller-Hinton agar plates by swabbing twice in order to ensure an even distribution of the inoculum. The surface of the medium was then allowed to dry for 3–5 min (Tendencia, 2004), and a sterile forceps was used to place the antibiotic discs onto the surface of the agar to ensure complete contact between the disk and the agar surface. The plates were incubated at 37°C for 24 h, and the zone of inhibition was measured in millimeters by using a transparent meter ruler. The isolates that showed resistance to three or more antibiotics were considered as MDR isolates.
The multiple antibiotic resistance (MAR) index was determined for each isolate by using the following formula: MAR = a/b, where a represents the number of antibiotics to which the tested isolate showed resistance and b represents the total number of antibiotics to which the tested isolate was evaluated for susceptibility (Sandhu et al., 2016).
Plasmid profiling
Plasmid profiling was performed on selected MDR bacteria by screening them for the presence of extrachromosomal DNA using the method of Ezeamagu et al. (2017). Plasmids might be responsible for the resistance traits of the bacterial isolates to multiple antibiotics. Plasmid extraction was performed at the Molecular Epidemiology Unit of the Molecular Biology and Biotechnology Department, Nigerian Institute of Medical Research, Yaba, Lagos state, Nigeria, by using the TENS method. A 24-h-old broth culture of the MDR bacteria was transferred to Eppendorf tubes and centrifuged for 2 min at 13 000 rpm. The supernatant was decanted and then vortexed to resuspend the cells in 300 μl of TENS buffer (Tris 25 mM, EDTA 10 mM, NaOH 0.1 N, and SDS 0.5%). The solution was mixed by inverting the tube for 3–5 min until the solution became viscous. A volume of 150 μl of 3 M sodium acetate was added, and the solution was vortexed and centrifuged for 5 min in a microcentrifuge to pellet cell debris and chromosomal DNA. The supernatants were transferred to fresh Eppendorf tubes by decanting, and 900 μl of ice-cold absolute ethanol was added. The sample was vortexed for another 10 min to pellet plasmid DNA. The supernatants were discarded, and the pellet was washed twice with 1 ml of 70% ethanol and dried. The pellet was re-suspended in 40 μl of distilled water. The extracted plasmid was resolved by 0.8% agarose gel electrophoresis, and the purified plasmid DNA was stored at 4°C until further analysis.
Plasmid curing
A 24-h-old bacterial cell culture was transferred to 5 ml nutrient broth supplemented with 1 mg/ml acridine orange and incubated at 37°C for 48 h to 1 week. The cured organisms were then plated out on nutrient agar, and an antibiotic sensitivity test was performed twice (Silhavvy et al., 1984).
Results
A total of 81 bacterial isolates were obtained from the hawked soymilk samples collected from the Ibadan Polytechnic Community and were identified as Citrobacter spp. (52), Enterobacter (5), Escherichia coli (5), Shigella sp. (2), Serratia sp. (5), Proteus spp. (5), and Klebsiella sp. (6). Table 1 shows the results of the microbial load of the different samples of soymilk. The results revealed that the highest total heterotrophic bacterial count of 9.3 × 103 CFU/ml was recorded at Polytechnic North and South, while the least heterotrophic count of 3.2 × 107 CFU/ml was recorded at Polytechnic Central. The highest enteric bacterial count of 1.3 × 103 CFU/ml was recorded at Polytechnic Central, while the least enteric bacterial count of 1.7 × 107 CFU/ml was recorded at Polytechnic North.
Table 1
Total microbial counts of the hawked soymilk samples collected from the Ibadan Polytechnic Community
Biochemical characterization and identification of the bacterial isolates obtained from the hawked soymilk samples
Table 2 shows the probable identity of the 81 bacterial isolates obtained from the hawked soymilk samples from all axis of Ibadan Polytechnic according to the results of the biochemical tests and with reference to Bergey’s Manual of Systematic Bacteriology.
Table 2
Biochemical characterization and probable identity of bacteria isolated from the hawked soymilk samples collected from the Ibadan Polytechnic Community
Isolates S1–S13 represent Enterobacteriaceae isolates obtained from the hawked soymilk samples from Polytechnic North. They include Citrobacter spp. (C1, C2, C3, C4, C5, C6, C7, C8), Klebsiella spp. (K1, K2), Serratia spp. (S1, S2), and Shigella spp. (Sh1).
Isolates M1–19 represent Enterobacteriaceae isolates obtained from the hawked soymilk samples from Polytechnic South. They include Citrobacter spp. (C9, C10, C11, C12, C13, C14, C15, C16, C17, C18, C19, C20), Klebsiella spp. (K3), Serratia spp. (S3), Enterobacter (En1, En2), and Proteus spp. (P1, P2, P3).
Isolates G1–G21 represent Enterobacteriaceae isolates obtained from the hawked soymilk samples from Polytechnic East. They include Citrobacter spp. (C21, C22, C23, C24, C25, C26, C27, C28, C29, C30, C31, C32, C33, C34, C35), Serratia spp. (S4, S5), Klebsiella spp. (K4), Proteus spp. (P4), and Enterobacter spp. (En3, En4).
Isolates O1–O5 represent Enterobacteriaceae isolated from the hawked soymilk samples from Polytechnic West. They include Citrobacter spp. (C36, C37, C38, C39, C40, C41, C42, C43, C44, C45, C46), Salmonella spp. (Sa1, Sa2), Shigella spp. (Sh2), and Klebsiella spp. (K5).
Isolates B1–B13 represent Enterobacteriaceae isolated from the hawked soymilk samples from Polytechnic Central. They include Citrobacter spp. (C47, C48, C49, C50, C51, C52), Enterobacter spp. (En5, E. coli (E1, E2, E3, E4, E5), and Klebsiella spp. (K6).
Antibiotic susceptibility pattern of Enterobacteriaceae isolates from the hawked soymilk samples
Antibiotic resistance of Enterobacteriaceae isolates from the North axis
Table 3A shows the antibiotic sensitivity pattern of the Enterobacteriaceae isolates from the hawked soymilk samples from the Polytechnic North axis. All isolates were susceptible to all the antibiotics used, except isolate C8 (Citrobacter spp.). Isolate C8 was resistant to ertapenam, gentamicin, aztreonam, amoxicillin-clavulanate, ciprofloxacin, ceftazidime, trimethoprim-sulfamethoxazole, and cefpodoxime with inhibition zones of 14–17 mm, 18–20 mm, 18–20 mm, 18–20 mm, 13–14 mm, 16–20 mm, 11–15 mm, and 10–14 mm, respectively.
Table 3A
Antibiotic sensitivity pattern of Enterobacteriaceae isolates from hawked soymilk samples from the Ibadan Polytechnic North Axis
[i] ETP – ertapenem (10 μg), CN – gentamicin (10 μg), ATM – aztreonam (30 μg ), AMC – amoxicillin-clavulanate (20/10 μg), CIP – ciprofloxacin (5 μg), CAZ – ceftazidime (30 μg), SXT – trimethoprim-sulfamethoxazole (1.25/23.75 μg), CPD – cefpodoxime (10 μg), S – susceptibility, I – intermediate, R – resistance
Antibiotic resistance of Enterobacteriaceae isolates from the South axis
Table 3B shows the antibiotic sensitivity pattern of the Enterobacteriaceae isolates from the hawked soymilk samples from the Polytechnic South axis. The tests revealed that all the isolates M1 to M19 were susceptible to almost all the antibiotics used. Isolates M2, M11, and M12 were resistant to amoxicillin-clavulanate, and M18 was resistant to ertapenem but susceptible to all the other antibiotics. Isolate M6 was resistant to ertapenem and cefpodoxime but sensitive to the other antibiotics.
Table 3B
Antibiotic sensitivity pattern of Enterobacteriaceae isolates from hawked soymilk samples from the Ibadan Polytechnic South Axis
Antibiotic resistance of Enterobacteriaceae isolates from the East axis
Table 3C shows the antibiotic sensitivity pattern of the Enterobacteriaceae isolates from the hawked soymilk samples from the Polytechnic East axis. Isolates G1, G4, G5, G6, G8, G9, G12, and G16 to G20 were susceptible to almost all the antibiotics used. Isolates G3, G11, G13, G14, G15, and G21 were resistant to one of the antibiotics, while G2 and G7 were resistant to two of the tested antibiotics. Isolate G10 was resistant to three antibiotics, including ertapenem, aztreonam, and cefpodoxime, and thus showed a multidrug resistance pattern.
Table 3C
Antibiotic sensitivity pattern of Enterobacteriaceae isolates from hawked soymilk samples from the Ibadan Polytechnic East Axis
Antibiotic resistance of Enterobacteriaceae isolates from the West axis
Table 3D shows the antibiotic susceptibility pattern of the Enterobacteriaceae isolates from the hawked soymilk samples from the Polytechnic West axis. Isolates O4, O6 to O8, O11, O13, and O15 were susceptible to almost all the antibiotics used. Isolates O3 and O9 were resistant to one antibiotic, while isolates O1, O2, O5, O10, and O12 were resistant to two antibiotics. Isolate O4 was resistant to aztreonam, ceftazidime, trimethoprim-sulfamethoxazole, and cefpodoxime, while isolate O14 was resistant amoxicillin-clavulanate, trimethoprim-sulfamethoxazole, and cefpodoxime; hence, isolates O4 and O14 showed a multidrug resistance trait.
Table 3D
Antibiotic sensitivity pattern of Enterobacteriaceae isolates from soymilk samples hawked in the Ibadan Polytechnic West Axis
Antibiotic resistance of Enterobacteriaceae isolates from the Central axis
Table 3E shows the antibiotic sensitivity pattern of the Enterobacteriaceae isolates from the hawked soymilk samples from the Polytechnic Central axis. Isolates B1 to B9 and B11 to B13 were susceptible to almost all the antibiotics used, while isolate B10 was resistant to three antibiotics (ertapenem, aztreonam, and cefpodoxime); hence, it showed a multidrug resistance trait.
Table 3E
Antibiotic sensitivity pattern of Enterobacteriaceae isolates from hawked soymilk samples from the Ibadan Polytechnic Central Axis
Susceptibility profile of isolates to antibiotics
Table 4 shows the results of percentage resistance and sensitivity of Enterobacteriaceae isolates from the hawked soymilk samples from all axis of the Polytechnic Community. A total of 88% of the Enterobacteriaceae isolates were more susceptible to trimethoprim-sulfamethoxazole, while 26% isolates were resistant to cefpodoxime. The isolates showed low resistance to ciprofloxacin (3%), while 47% of the isolates showed low susceptibility to cefpodoxime.
Table 4
Antibiotic susceptibility profile of isolates [%]
| Antibiotics | Inhibition [%] | Intermediate [%] | Susceptible [%] |
|---|---|---|---|
| ETP | 10.00 | 11.00 | 79.00 |
| CN | 6.00 | 8.00 | 86.00 |
| ATM | 15.00 | 10.00 | 75.00 |
| AMC | 13.00 | 17.00 | 70.00 |
| CIP | 3.00 | 44.00 | 53.00 |
| CAZ | 11.00 | 26.00 | 63.00 |
| SXT | 12.00 | 0.00 | 88.00 |
| CPD | 26.00 | 27.00 | 47.00 |
[i] ETP – ertapenem (10 μg), CN – gentamicin (10 μg), ATM – aztreonam (30 μg ), AMC – amoxicillin-clavulanate (20/10 μg), CIP – ciprofloxacin (5 μg), CAZ – ceftazidime (30 μg), SXT – trimethoprim-sulfamethoxazole (1.25/23.75 μg), CPD – cefpodoxime (10 μg), S – susceptibility, I – intermediate, R – resistance
Table 5 shows the results of the multidrug resistance pattern of the Enteriobacteriaceae isolates from the hawked soymilk samples. Soymilk samples from the Polytechnic West axis had the highest occurrence of the MDR Enterobacteriaceae isolates with isolates O4 (Shigella spp.) and O14 (Klebsiella spp.) showing 50% and 37.5% resistance, respectively, to the antibiotics tested. Isolate S13 (Citrobacter spp.) from the Polytechnic North axis showed 62.5% resistance, while isolate B10 from the Polytechnic Central axis (E. coli ) showed 37.5% resistance to the antibiotics tested. Isolate G10 (Proteus spp.) from the Polytechnic East axis exhibited 37.5% resistance to all the antibiotics used.
Table 5
Multidrug resistance pattern of Enterobacteriaceae isolates from hawked soymilk samples from the Ibadan Polytechnic Community
Table 6 shows the results of the MAR index of the Enterobacteriaceae isolates from the hawked soymilk samples. The MAR index of the isolates ranged from 0.4 to 0.6. Isolate S13 (Citrobacter spp.) showed the highest MAR index of 0.6, isolate O4 (Shigella spp.) had an MAR index of 0.5, and isolates G10, O14, and B10 had an MAR index of 0.4.
Table 6
MAR index values of Enterobacteriaceae isolates from hawked soymilk samples from the Ibadan Polytechnic Community
| Isolates | CN | AMC | CIP | CAZ | SXT | CPD | ATM | ETP | MAR index |
|---|---|---|---|---|---|---|---|---|---|
| S13 | S | R | R | R | R | R | S | S | 0.6 |
| G10 | S | S | I | I | S | R | R | R | 0.4 |
| O4 | S | I | S | R | R | R | R | S | 0.5 |
| O14 | S | R | I | I | R | R | S | S | 0.4 |
| B10 | S | I | I | I | S | R | R | R | 0.4 |
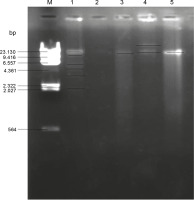
The MDR enteric bacteria were then screened for the plasmids they carried. Figure 1 shows the gel electrophoresis plasmid profile of the MDR isolates. The plasmids obtained from the MDR enteric bacteria had different molecular weights, with the maximum molecular weight of 17.2 kbp. Isolate O14 (Table 7) (Klebsiella spp.) had the highest number of plasmid bands, with 6 plasmid bands of molecular weights ranging from 2.7 to 12.9 kbp, and isolate G10 (Proteus spp.) was found to have lost its plasmid. One of the plasmid bands of isolate O14 (Klebsiella spp.) had the same plasmid size of 11 kbp as that of isolates S13, B10, and O4 (Citrobacter spp., E. coli, and Shigella spp., respectively) (Table 7). Isolate B10 was found to harbor 2 plasmid bands with molecular weights of 17.2 and 11.2 kbp, while isolate S13 and 04 had a single plasmid band with molecular weight of 11.2 kbp (Table 7).
Table 7
Number of plasmids with their molecular weight and mobility of the multidrug-resistant isolates
[i] Dye font (df) – 55 mm, correlation coefficient (r) – 0.9615 (calculation obtained from nsilico.ehu.es/mini_tools/molecular_weight/index.php)
Fig. 1
Gel electrophoresis for plasmid profiling of multidrug resistant Enterobacteriaceae : lane M – DNA–Hin d III Digest, lane 1 – isolate O14 (Klebsiella spp.), lane 2 – isolate G10 (Proteus spp.), lane 3 – isolate S13 (Citrobacter spp.), lane 4 – isolate B10 (E. coli), lane 5 – isolate O4 (Shigella spp.)
To verify whether the resistance to antibiotics is carried on plasmids, curing was performed to eliminate the plasmids from bacteria. After curing, the antibiotic sensitivity tests for the MDR bacteria were repeated. Table 8 shows the antibiotic sensitivity patterns of the MDR Enterobacteriaceae isolates before and after the elimination of plasmids (curing) from the bacteria. Before curing, isolate O14 (Klebsiella spp.) showed resistance to amoxicillin-clavulanate, trimethoprim-sulfamethoxazole, and cefpodoxime, and even after curing, the isolate was found to show the same resistance pattern. Isolate S13 (Citrobacter spp.) was resistant to amoxicillin-clavulanate, ciprofloxacin, ceftazidime, trimethoprim-sulfamethoxazole, and cefpodoxime; however, after curing, it became sensitive to ciprofloxacin, ceftazidime, trimethoprim-sulfamethoxazole, and cefpodoxime, but remained resistant to amoxicillin-clavulanate. Isolate B10 (E. coli ) showed resistance to cefpodoxime, aztreonam, and ertapenem, and even after curing, the resistance pattern remained the same. Isolate O4 (Shigella spp.) was resistant to trimethoprim-sulfamethoxazole, cefpodoxime, aztreonam, and ceftazidime; however, after curing, it became susceptible to trimethoprim-sulfamethoxazole and cefpodoxime, but remained resistant to aztreonam and ceftazidime.
Table 8
Antibiotic sensitivity pattern before and after curing of plasmids of the multidrug-resistant Enterobacteriaceae
| Isolate code | AMC | CIP | CAZ | SXT | CPD | ATM | ETP |
|---|---|---|---|---|---|---|---|
| O14 B/c A/c | R R | – | – | R R | R R | – | – |
| S13 B/c A/c | R R | R S | R S | R S | R S | – | – |
| B10 B/c A/c | – | – | – | – | R R | R R | R R |
| O4 B/c A/c | – | – | R I | R S | R R | R R | – |
[i] ETP – ertapenem (10 μg), ATM – aztreonam (30 μg), AMC – amoxicillin-clavulanate (20/10 μg), CIP – ciprofloxacin (5 μg), CAZ – ceftazidime (30 μg), SXT – trimethoprim-sulfamethoxazole (1.25/23.75 μg), CPD – cefpodoxime (10 μg), B/c – before plasmid curing, A/c – after plasmid curing, R – resistance, I – intermediate, S – susceptible
Discussion
The present work was designed to determine the occurrence and plasmid profiling of MDR Enterobacteriaceae isolates obtained from soymilk hawked in the Ibadan Polytechnic Community. The microbial load of different samples of soymilk was very high, which indicated a high level of contamination of the soymilk samples with Enterobacteriaceae. This result agreed with the earlier reports of Adeleke et al. (2000), Schlegelova et al. (2002), Guta et al. (2002), and Okpalugo et al. (2008) on the evaluation of the occurrence of pathogenic microorganisms in soymilk. The high microbial load of Enterobacteriaceae observed in the soymilk samples in this study might have emanated from poor handling, use of contaminated raw materials, unhygienic processing environment, low literacy level, and lack of good manufacturing practices by producers. Mbajiuka et al. (2014) also reported the occurrence of microorganisms in soymilk; these microorganisms can survive in soymilk milk possibly due to the presence of rich source of proteins, fats, and minerals. The presence of proteolytic enzymes in the bacteria confers them with the ability to catabolize soymilk into simple end products that are important for their growth and metabolism (Gandi, 2009). In the present study, Citrobacter spp. were found to be predominant species in soymilk samples, and this finding differed from the report of Liamngee et al. (2013) who showed a high occurrence of E. coli and Klebsiella spp. in the soymilk sample sold in Makurdi metropolis, Nigeria.
The high susceptibility of Enterobacteriaceae isolated from hawked soymilk to Trimethoprim-sulfamethoxazole was reported previously (Osterblad et al., 1999; Oliver et al., 2002). The ability of bacteria to demonstrate antimicrobial resistance might be due to the acquisition of plasmid-encoded β-lactamases, which could be broad-spectrum, narrow-spectrum, or extended-spectrum β lactamases (ESBLs) (Mulla et al., 2012). In the current study, 6% of the Enterobacteriaceae isolates were resistant to gentamicin. This observation is similar to the findings of Yulistiani et al. (2017) who reported that 5.9% of the total Enterobacteriaceae strains tested were resistant to gentamicin. Patel et al. (2008) reported that 15% of the total number of Enterobacteriaceae strains were resistant to Aztreonam, which is in agreement with the findings of this study. The resistance of bacteria to Aztreonam could be due to the inability of this drug to bind to the penicillin-binding proteins, thereby making it ineffective against them. As observed in this study, the resistance of Enterobacteriaceae to amoxicillin-clavulanate did not agree with the report of Mulla et al. (2011) who showed a high sensitivity of Enterobacteriaceae to amoxicillin-clavulanate. Citrobacter spp. showed the highest percentage of multidrug resistance, which could be due to the worldwide prevalence of ESBLs-producing Citrobacter strains, which have been earlier reported by Park et al. (2005), Moland et al. (2006), and Choi et al. (2007). The presence of quinolone-resistant genes on plasmids could also be responsible for this resistance (Park et al., 2007; Zhang et al., 2012). The loss of plasmids observed in a member of the isolated Enterobacteriaceae could be due to frequent subculturing and instability of plasmids. Akingbade et al. (2013) found that 10 of the 27 MDR Enterobacteriaceae strains analyzed carried plasmids, while the others had lost their plasmids. However, Ineta et al. (2018) reported that 70% of the Enterobacteriaceae isolates were devoid of plasmids, while the remaining 30% carried plasmids with molecular weights of 30 kbp and above. The presence of two plasmid bands in E. coli as observed in the present study agrees with the earlier report of Rabee et al. (2016) who observed the occurrence of multiple plasmids of different sizes in E. coli, which could be responsible for its high antimicrobial resistance (Alsultan et al., 2013; Alanazi et al., 2018) and display of resistance after curing (Suhani et al., 2017). In the bacteria that maintained their resistance after curing, the acquisition of resistance did not seem to be plasmid-mediated. However, some of the MDR bacteria were found to be partly resistant and partly sensitive to the antibiotics after curing. This indicates that the resistance might be mediated chromosomally (Reboucas et al., 2011; Letchumanan et al., 2015).
Conclusion
Soymilk sold in the sampled areas was highly contaminated with Enterobacteriaceae, thus indicating poor hygiene standards of the production process. The presence of coliform bacteria in soymilk indicates a high level of contamination. Hence, proper monitoring by food safety authorities and strict adherence to aseptic conditions should be followed during the production of soymilk. The present study also revealed that plasmids might confer antibiotic resistance to bacterial strains that are currently susceptible to drugs.










