Introduction
To produce recombinant proteins in large amounts, suitable plants can be used as a convenient alternative to mammalian species, insects, yeast cell cultures, and microbial industrial bioreactors. Tobacco is characterized by a high production of biomass (almost 170 tons per hectare), high seed yield, self-pollination, maternal chloroplast inheritance, and relatively easy genetic engineering as compared to other plants; thus, it constitutes a good option for molecular farming and production of pharmaceutical recombinant proteins (Ivanov et al., 2020). Compared to other systems, the production of pharmaceutical proteins through plant chloroplast genetic engineering has many advantages such as a high level of recombinant protein accumulation in chloroplasts due to their polyploidy nature, limited number of protein degradation pathways, lack of side effects, and silencing because of homologous directional integration of transgene in specific regions in the chloroplast genome and maternal heritage of chloroplast genes (Abd-Aziz et al., 2020; Kopertekh and Schiemann, 2019). Furthermore, the prokaryotic expression system of the chloroplasts enables researchers to introduce multiple genes within an operon to express an entire metabolic pathway (Schindel et al., 2018).
Interferon-beta (IFN-β) is a member of type I interferons (IFNs). It is known for its antiviral and immunomodulatory characteristics (Morowvat et al., 2014). IFN-β is one of the main therapies for treating multiple sclerosis (MS), although many patients cannot afford this treatment because of its high cost (Thompson et al., 2013). Because of the clinical success of IFN-β, there is a growing global demand for this important product (McCrae, 2021; Palesh et al., 2008; Wood, 2020). To respond to this increasing demand, a cheaper production system is required to be developed. The production of pharmaceutical recombinant proteins using transgenic plants is an economical and effective method. Compared to the expression systems for recombinant proteins using nuclear transformation systems, the chloroplast expression system offers several advantages, including high expression level, occurrence of the position effect, maternal inheritance that lessens the risk of dispersal of the transgene into the environment, ability to express polycistronic genes, and no obvious gene silencing (Karimi et al., 2013; Sheikh-Mohamadi et al., 2017; Sheikh Mohamadi et al., 2018; Sheikh Beig Goharrizi et al., 2016). Svab et al. (1990) developed a stable transformation protocol for tobacco chloroplasts by using the gene gun method. In the past two decades, remarkable progression has been made in producing valuable recombinant proteins using plant chloroplast expression systems (Adem et al., 2017), for example, the production of monoclonal antibody (Fischer et al., 2004), stable antibiotic protein (Oey et al., 2009), interferon-alpha (Arlen et al., 2007), and recombinant single-domain monoclonal antibody (Rajabi-Memari et al., 2006).
Model plants exhibit several properties such as easy transformability, short life cycle, high seed production, and simple propagation by seeds, which make them appropriate for use in functional genomics (Dejahang et al., 2018; Farajpour et al., 2011). Several methods are available to transform chloroplasts, such as transformation by polyethylene glycol and gene gun methods. Polyethylene glycol is used for plant species with chloroplasts that can be extracted and regenerated (Bock, 2014). Simplicity and the ability to transform different plant species make gene gun transformation a convenient and popular method (Adem et al., 2017). It has been assumed that the engineering of tobacco chloroplasts is an efficient and cost-effective method to produce IFN-β. Therefore, the present study aimed to produce recombinant human interferon-beta (rhIFN-β) by expression in tobacco chloroplasts.
Materials and methods
Plant materials and growth conditions
Green leaves of Nicotiana tabacum L. cv. “Samsun” were used for chloroplast transformation. The tobacco seeds were obtained from the National Plant Gene-bank, Karaj, Iran. They were sterilized by 70% ethanol for 1 min, incubated in 2% sodium hypochlorite for 15 min, and cultured on MS medium containing 30 g and 8 g/l of sucrose and agar, respectively. The pH of the medium was adjusted to 5.6. The cultures were kept under 16 h photoperiod (40 μmol ∙ m–2 ∙ s–1) provided by 40-W white fluorescent lamps at 25 ± 2°C for 3–4 weeks. The green leaves were then transferred to a Revised Medium for Organogenesis of Nicotiana plumbaginifolia (RMOP) medium one day before gene bombardment (Scotti et al., 2009).
Plasmid preparation
The tobacco chloroplast codon-optimized ifn-β gene was developed by ShineGene Molecular Biotech Inc. (Shanghai, China). The ifn-β codon sequence was optimized by GenScript’s patented OptimumGeneTM algorithm. The pVSR326 chloroplast vector, which was kindly provided by Dr. Vanga Siva Reddy (International Centre for Genetic Engineering and Biotechnology, New Delhi, India), was used for plastid transformation. The vector is capable of integrating the desired foreign genes into the tobacco chloroplast genome by a site-specific integration between the rbcL and ccD genes (nucleotides 58 056–60 627). The Nco I and Sac I restriction enzymes were used for inserting the subclone ifn-β coding sequence into the pVSR326 vector according to a method described by Reddy et al. (2002). The integration of the ifn-β gene was verified by excising it using the abovementioned restriction enzymes and sequencing the obtained sequence. The recombinant plasmid was termed pVSR326-IFNβ. One positive clone was selected for further experiments (Karimi et al., 2013). The aadA gene was used as a selectable marker that provided spectinomycin resistance. The rice psbA promoter and terminator were used in the recombinant pVSR326-IFN-β plasmid to control the expression of the ifn-β gene (Reddy et al., 2002). The GGAGTC ribosome binding site was used for enhancing the translation upstream of the ifn-β coding sequence. The QIAGEN Plasmid Midi Kit (QIAGEN, Germany) was used for the isolation of plasmid DNA according to the manufacturer’s protocol.
Plant transformation and regeneration
The PDS-1000He (Bio-Rad, Hercules, CA, USA) gene gun with 900 psi was used for the transformation of tobacco leaves through the gene bombardment method. It involved using the pVSR326-IFN-β plasmid DNA-coated gold (0.6 microns) micro-projectile (Daniell, 1997). Nearly 100 tobacco leaves were bombarded with the prepared plasmid. Another 100 tobacco leaves were transformed with the native Agrobacterium tumefaciens without any constructs as control for bioactivity assay.
After bombardment, the leaves were placed on the RMOP medium and incubated in dark at 25°C for 48 h under a 16 : 8-h photoperiod. The bombarded leaves were cut into 5 × 5 mm pieces and cultured in RMOP medium containing NAA (0.1 mg/l), BA (1 mg/l), and spectinomycin dihydrochloride (500 mg/l). They were left until a suitable time for the selection and regeneration of transplastomic tobacco cells. Six to eight weeks later, putative transplastomic shoots were identified as resistant to spectinomycin. Three rounds of regeneration involved using small sections of the shoots as explants on the RMOP medium containing spectinomycin. The antibiotic-resistant shoots were further cultured on the basic MS medium in the growth chamber with the 16 : 8-h light/dark photoperiod at 25°C. The rooted transplastomic plants were potted and continued to grow in a growth room at 26°C and a 16 : 8-h photoperiod.
PCR analysis of transplastomic plants
DNA was extracted from the leaves of putative transplastomic plants and their wild counterparts by an i-genomic DNA extraction mini kit (iNtRON Biotechnology, Korea) according to the manufacturer’s protocol. A PCR analysis was performed to confirm the integration of the ifn-β and aadA genes into the chloroplast genome. The ifn-β forward (5′– CGACAAGATTCTTCTTCTACTGGATGG–3′) and reverse primers (5′– GTCGACATTACGAAGATAACCTGTCA–3′) were used for PCR analysis. The following forward (5′–GGATCCATGGCTCGTGAAGCGGT–3′) and reverse (5′–CCATGGATCCCTCCCTTTATTTGCCAACTACCT–3′) primers were used for ensuring the integration of aadA in transplastomic plants.
The PCR reaction mixture contained 1 × PCR buffer, 0.2 mM dNTP mixture, 1 μM of each primer, one unit of Taq DNA polymerase, and 1 μl digested DNA in a total volume of 20 μl. The following PCR conditions were used: 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 45 s at 58°C, and 1 min 72°C, and a final extension of 5 min at 72°C. The PCR products were then separated by electrophoresis on a 1% agarose gel. The gels were stained with ethidium bromide and visualized by ultraviolet illumination.
Southern blot analysis
Southern blot analysis was performed to analyze the integration of the ifn-β gene into the tobacco chloroplast genome. Twenty micrograms of genomic DNA from the transplastomic and non-transplastomic plants were digested by 10 units of Cla I in CutSmart buffer (New England BioLabs, USA) at 37°C overnight. The products were separated by electrophoresis on a 1% agarose gel and were blotted on the Hybond N membrane (Amersham, Buckinghamshire, UK) according to the standard procedure described by Sambrook Russel (2001). The detection of the ifn-β cassette was carried out by a 453-bp digoxigenin (DIG)-labeled probe (P1) that was produced by the PCR DIG Labeling MIX (Roche, Germany) using ifn-β as a template and specific forward (5′–CGACAAGATTCTTCTTCTACTGGATGG–3′) and reverse (5′–GTCGACATTACGAAGATAACCTGTCA–3′) primers following the manufacturer’s instructions.
The homoplasty of transplastomic plants was confirmed by an 820-bp DIG-labeled rbcl probe (P2) using the following specific forward (5′–CTCACAACCATTTATGCG–3′) and reverse (5′–GTGGACGTTTTGGATAAG–3′) primers and tobacco DNA as a template according to the procedure described by Karimi et al. (2013). Hybridization and detection were performed according to the manufacturer’s instructions (Roche).
Transcriptional and translational analyses
RT-PCR was performed for analyzing the transcription of the ifn-β gene. In the first step, total RNA was isolated from the transplastomic and wild-type plant leaves using the RNeasy Mini Kit (QIAGEN, Germany) according to the manufacturer’s protocol. The RT-PCR reaction was performed in a final volume of 20 μl containing the gDNA Wipeout Buffer, RNase-free water, Quantiscript Reverse Transcriptase, RT primer mix, Quantiscript RT Buffer, and 1 μg of total RNA. The reaction was performed in an MJ Research Thermal Cycler under the following conditions: 2 min at 42°C for gDNA elimination, immediately followed by placing the reaction tube on ice for adding the reaction mixture, 30 min at 42°C for reverse transcription, and 3 min at 95°C for enzyme inactivation. The presence and expression level of ifn-β were determined by the RT-PCR analysis. The RT-PCR conditions and primers were the same as mentioned above, except that cDNA was used instead of DNA template. The RT-PCR product was separated by electrophoresis on a 1% agarose gel and then stained with 0.5 mg/l ethidium bromide. The PCR products were visualized under UV illumination. A 1-kb DNA ladder (Fermentas) was used as the size marker.
SDS-PAGE and western blot analyses were used to confirm translation of the ifn-β gene. The isolation of total proteins from the leaves, together with SDS-PAGE and immunoblotting procedures, were performed as described previously by Karimi et al. (2013). Fifty micrograms of TSP was separated on a 10% SDS-PAGE. The bands were transferred to a nitrocellulose membrane (Roche) by using a Mini Trans-Blot electrophoretic transfer cell (BioRad, USA) in transfer buffer (50 mM Tris base, 40 mM glycine, 0.04% SDS, and 20% methanol, pH 8.3). The membrane was washed three times in PBST buffer, and the color was developed using a horseradish peroxidase (HRP) staining solution (DAB; Sigma). The chromogenic reaction was stopped by rinsing the membrane twice with distilled water.
The expression of the ifn-β recombinant gene was quantified in transplastomic plants by ELISA. Fully grown leaves from transplastomic and wild-type plants were ground with liquid nitrogen and homogenized in PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4). An ELISA test was performed using commercial anti-IFN-β antibodies (Sigma), where the recombinant IFN-β (Sigma, St. Louis, MO, USA) served as the standard as described previously (Wirth et al. 2004).
Leaf extract preparation
For the leaf extract preparation, fresh leaves were air-dried for 5 days at 30°C, and 100 g of dried material was macerated in 2 l of distilled water. The extract was decanted after 24 h. The filtrate was dried in an oven at 40°C for 4 days. The dried extract was weighed and dissolved in distilled water to obtain a 200 mg/ml stock concentration.
Bioactivity assay of rhIFN-β
The bioactivity assay for rhIFN-β was performed based on cytoplastic inhibition of vesicular stomatitis virus (VSV) in human amnionic cells (WISH), as described by Langford et al. (1981). Human amnionic cells were seeded onto a 96-well tissue culture plate and incubated at 37°C overnight. After a confluent monolayer of cells was formed, approximately 100 μl of the hIFN-β standard (Sigma-Aldrich, Germany) was added to the wells along with the addition of leaf extracts of the rhIFN-β that had been purified. The extract of leaves infiltrated with native A. tumefaciens without any constructs was considered as the negative control, whereas the standard hIFN-β was used as the positive control. Serial half-log10 dilutions (10–0.5) of protein samples were used. Each sample was assayed in three replicates. The plate was incubated at 37°C for 16 h. The medium was then disposed of, and the cells were washed twice with sterile phosphate-buffered saline (PBS). Next, 100 μl (100 TCID50) VSV was inoculated into each well, and the plate was incubated at 37°C. Virus infection was visually checked, and the plate was incubated for 30 h until all cells that had not received the IFN-β treatment were infected by VSV. The IFN-β activity was measured in international units (IU) per mg of TSP of tobacco leaf according to the method of Langford et al. (1981). One unit of IF is present in the dilution of a sample that inhibits the formation of plaques equal to 50% of that in the virus control.
Results
Plant transformation and regeneration
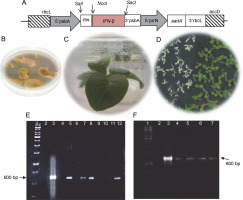
The tobacco chloroplast codon-optimized ifn-β gene sequence (509 bp) was deposited in the GeneBank (MH285815.1). The recombinant gene contained an ifn-β1a coding sequence, lacking 21 amino acids from the protein N-terminus. The rice psbA promoter and terminator were used as transcriptional elements in pVSR326-IFN-β plasmids, while the aadA gene was used as a selectable marker that provided spectinomycin resistance. The ribosome binding site 5′–GGAGTC–3′ was used for enhancing the translation upstream of the ifn-β coding sequence. The rbcL-accD flanking sequences integrated the transgene cassette (6.4 kb) into the chloroplast genome between the rbcL-accD region through two homologous recombination (Fig. 1A) (Reddy et al., 2002). The 25 antibiotic-resistant green calli were obtained from approximately 100 tobacco leaves that were bombarded by the DNA of pVSR326-IFN-β. The antibiotic-resistant calli were regenerated on an RMOP medium (Fig. 1B and 1C). The transplastomic lines were regenerated four times on the selection medium, and after 8 months, they were transferred to MS medium containing 500 mg/l of spectinomycin, so that rooting occurred before the transfer to a growth chamber. All 25 antibiotic-resistant green calli were successfully regenerated and rooted on MS medium containing 500 mg/l spectinomycin. The seeds of the regenerated plants were cultured on a medium containing spectinomycin and were assessed for stability regarding the integration of ifn-β in transplastomic plants (Fig. 1D).
Fig. 1
A) Schematic structure of the recombinant ifn-β gene in the pVSR326 chloroplast transformation vector: the aadA spectinomycin resistance gene was used as a selectable marker; 5′prrn rice 16SrRNA operon promoter; Rbs ribosome binding site; B) spectinomycin-resistant green calli; C) transplastomic regenerated plant; D) germination of transplastomic T1 (green) and wild-type (white) seeds; E) PCR results of the putative transplastomic plants with ifn-β primers (arrows); F) detection of putative transplastomic plants using aadA primers
PCR and Southern blot analyses of transplastomic plants
PCR using the ifn-β and aadA specific primers was performed for 25 spectinomycin-resistant shoots, among which 4 showed both the 509 and 800 bp bands, representing the integration of the ifn-β and aadA genes, respectively (Fig. 1E and 1F).
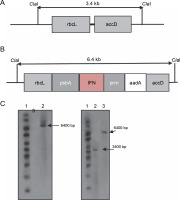
To verify the plant homoplasmy and the correct integration of the cassette between the rbcl and accD genes, DNA blot analysis was performed on PCR-positive transplastomic lines. In the chloroplast genome, the ClaI enzyme was used to cut the 5′ and 3′ borders of rbcl and accD sequences, respectively (Fig. 2A and 2B). The digestion of chloroplast DNA with the Cla I enzyme formed a 6.4-kb hybridizing band by a DIG-labeled ifn-β probe, which confirmed the insertion of ifn-β into the transplastomic chloroplast genome. The wild-type plants showed no detectable band (Fig. 2C). Homoplasmy was confirmed using the rbcL probe, which identified the 6.4 and 3.4 kb bands in transplastomic and wild-type plants, respectively (Fig. 2D).
Fig. 2
Southern blot analysis of the tobacco transplastomic line. A) graphic presentation of the Cla I restriction scheme in the wild-type chloroplast genome; B) graphic presentation of the Cla I restriction scheme in the transplastomic tobacco chloroplast genome; C) lane 1 – 1-kb DNA ladder, lane 2 – non-transplastomic control plant DNA, lane 3 – transplastomic line; the ifn-β probe was hybridized to a 6.4-kb fragment in the transplastomic line; D) lane 1 – 1-kb DNA ladder, lane 2 – wild-type plant DNA sample, lane 3 – plant DNA sample of the transplastomic line; the rbcL probe hybridized to a 7.4-kb fragment in the transplastomic line and to a 3.4-kb fragment in the wild-type plant
Transcription and translation analyses of transplastomic plants
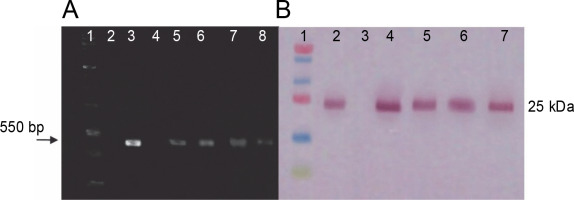
RT-PCR and western blot assays were performed to identify the transcription and translation products of ifn-β in transplastomic plants. RT-PCR results were obtained using ifn-β-specific primers and showed an approximately 550 bp ifn-β mRNA, thereby confirming the transcription of the ifn-β gene in the chloroplasts of transplastomic lines, whereas no band was detected in wild-type plants (Fig. 3A). Western blotting assay with the IFN-β-specific antibody detected the IFN-β protein. The blotting assay showed an approximately 25 kDa band in the transplastomic plants, indicating the production of the IFN-β protein in the chloroplast of these plants, while no bands were observed in the wild-type plants (Fig. 3B).
Fig. 3
RT-PCR and western blotting assay results. A) RT-PCR analysis performed on leaves of transplastomic lines and wildtype plants by using ifn-β-specific primers; lane 1 – PCR product without cDNA template (negative control), lane 2 – 1-kb DNA ladder (Fermentas), lane 3 – total RNA as the template, lane 4 – PCR product from pVSR326-IFNβ DNA as the template (positive control), lane 5 – DNA of the wild-type plant, lanes 6–9 – transplastomic lines; B) the IFN-β protein blotting assay in transplastomic lines by using anti-IFN-β antibody: lane 1 – protein molecular weight marker (Fermentas), lane 2 – positive control, lane 3 – 20 μg of TSP from wild-type plants, lane 4–7 – 20 μg TSP from transplastomic lines
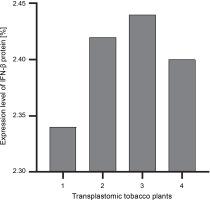
The ELISA assay was used to quantify the IFN-β protein in transplastomic plants. The absorbance readings of the samples were compared using the diluted grades of the IFN-β standard antigen. The IFN-β TSP percentage was calculated using the Bradford method (Sambrook and Russel, 2001). The percentage of expressed IFN-β was calculated as the percentage of TSP by using the Bradford assay. The amount of the IFN-β protein was approximately 2.4–2.5% of TSP (Fig. 4).
Bioactivity assay of rhIFN-β
The wells with sample cells and the standard hIFN-β showed normal growth after 30 h of infection with the VSV, while the control wells containing the extract of leaves with native A. tumefaciens without any constructs exhibited severe pathological changes as they became necrotic and did not show normal growth. By using hIFN-β as the standard, the bioactivity of the recombinant rhIFN-β was measured to be 2.9 × 104 IU/mg of TSP. The bioactivity assay showed that the rhIFN-β could protect human amnionic cells against VSV.
Discussion
Plastid genetic engineering in plants has significantly progressed during the past decades and has become a promising method for the production of high-value metabolites and proteins, e.g., industrial enzymes, biopharmaceuticals, and diagnostic reagents (Clarke et al., 2013; De Marchis et al., 2012; van Eerde et al., 2019; Younessi-Hamzekhanlu et al., 2021). The diverse features of chloroplast transformation include the dynamic concept of site-specific integration of the transgene into the plastid genome (Limaye et al., 2006), lack of position effects and epigenetic gene silencing (Daniell et al., 2001; Rigano et al., 2013; Ruhlman et al., 2010), insertion of transgenes in operons (Lu et al., 2013), and the biosafety profile of transplastomic plants. These qualities make chloroplast transformation an appropriate method for use in plant biotechnology. Currently, many recombinant proteins are expressed in chloroplasts, and efforts have been made to produce different pharmaceutical proteins in plant chloroplast (Hoelscher et al., 2018; Razmi et al., 2019; van Eerde et al., 2019). In the present study, the codons of the ifn-β coding sequence were optimized for expression in tobacco chloroplasts (Krasniqi et al., 2018). The production of the rhIFN-β protein in tobacco chloroplasts was confirmed by protein blot analysis using IFN-β-specific antibodies, where, as expected, a protein of approximately 25 kDa was detected.
A high level of expression of foreign proteins in plastids is a major development in plant biotechnology. It makes plastid transformation as an ideal approach for large-scale production of recombinant proteins (Gao et al., 2012). It has been reported that the expression of the HIV entry inhibitor griffithsin in the tobacco protoplast exceeded 5% of the plant’s TSP (Hoelscher et al., 2018). A simple purification process of plastidic griffithsin from the leaves proved this to be a feasible method. Moreover, the protein was characterized by a high level of virus neutralization activity as compared to the activity of the protein expressed in bacteria (Hoelscher et al., 2018). The dried tobacco leaves also serve as a storable source material for large-scale production of the recombinant griffithsin protein (Hoelscher et al., 2018). Arlen et al. (2007) documented a high level (up to 20% of TSP) of IFN-α2b production in tobacco chloroplasts. The transgenic IFN-α2b showed similar in vitro and in vivo biological activities as compared to commercial products. Previous studies have reported different expression levels of recombinant proteins in the tobacco chloroplast, such as interferon alpha-5 at the level of 4,372 pg/g fresh weight (Khan and Nurjis, 2012) and Bacillus anthracis 83-kDa protective antigen (PA) at 18% of TSP (Watson et al., 2004). Our results showed that the expression level of the recombinant protein was relatively low, which was in agreement with earlier studies (Razmi et al., 2019; Schindel et al. 2018; Shen et al., 2010). Many studies have indicated that the low expression level of recombinant proteins in chloroplasts could be attributed to codon usage, translation efficiency, promoter, and protein stability in chloroplasts (Ahmad et al., 2020; Apel et al., 2010; De Marchis et al., 2012; Rowland et al., 2015; Soria-Guerra et al., 2009; Younessi-Hamzekhanlu et al., 2020). Huy Kim (2019) reported a low expression level (0.07% of TSP) of the porcine epidemic diarrhea antigen in tobacco, whereas fusion with a stable B subunit of the cholera toxin protein increased the expression level by up to 1.4% TSP. However, several researchers have claimed that the mechanism of protein instability is still unclear and the prediction of foreign protein stability in plastids was not possible (Hoelscher et al., 2018). Razmi et al. (2019) expressed ifn-γ in the tobacco chloroplast at 0.42% of the TSP, which was much higher than its expression in nuclear-transformed plants (0.03%). These results could be ascribed to the lack of post-translational modifications in chloroplasts. Although the human IFN-β protein has an N-glycosylation site, the IFN-β protein expressed in transplastomic plants had almost the same size as that expressed in Escherichia coli (Fig. 3); this is because N-glycosylation is not known to occur within chloroplasts (Daniell et al., 2009). Chloroplasts are capable of performing some post-translational modifications such as folding, formation of disulfide bonds, and lipid modification, but they do not support glycosylation (Razmi et al., 2019).
The bioassay analysis confirmed the high bioactivity (2.9 × 104 IU/ml) of rhIFN-β expressed in tobacco chloroplasts. The protein could protect human amnionic cells against VSV infection. Previous studies have shown the bioactivity of rhIFN-γ in protecting monkey Vero cells against VSV (Heidari-Japelaghi et al., 2020), which is in agreement with our results. Even though previous studies have confirmed the in vitro and in vivo biological activities of recombinant pharmaceutical proteins in chloroplasts, we propose future studies to examine the biological activity of IFN-β following its expression in tobacco chloroplasts. In the present study, the recombinant IFN-β protein was successfully expressed at a reasonable level in the tobacco chloroplast, which may lead to the cost-effective production of IFN-β.
Conclusion
In the present study, the tobacco chloroplast transformation method was used to produce human IFN-β, which is known for its antiviral and immunomodulatory activities. It can be successfully used in treating MS. The integration of the ifn-β gene with the chloroplast genome was confirmed by molecular techniques at the DNA, RNA, and protein levels. The bioassay confirmed that the recombinant IFN-β had relatively high bioactivity in tobacco chloroplasts, as revealed by its capability to protect the human amnionic cells against VSV infection. Future experiments will determine the effect of ifn gene expression.