Introduction
Candida auris is a fungus that is emerging as a nosocomial pathogen (Chybowska et al., 2020; Sathyapalan et al., 2021). Multidrug-resistant strains of C. auris have been reported in over 30 countries worldwide (Chowdhary et al., 2017; Rudramurthy et al., 2017; Adams et al., 2018; Ruiz-Gaitán et al., 2018; Chow et al., 2019; Ademe and Girma, 2020). Resistance to major antifungal drugs including fluconazole, terbinafine, amphotericin B, and echinocandins has been reported in C. auris (Lamoth and Kontoyiannis, 2018; Chybowska et al., 2020). Around 40% of isolates are resistant to at least two antifungals, and 4–10% of isolates are resistant to most antifungals (Lockhart et al., 2017; Lamoth and Kontoyiannis, 2018).
C. auris can infect and colonize humans, especially in hospital settings (Belkin et al., 2018; Dahiya et al., 2020). These nosocomial infections are mostly associated with bacterial biofilm formation (Lamoth and Kontoyiannis, 2018) and can be transmitted between patients in healthcare settings, where healthcare personnel could also play a role in their transmission (Forsberg et al., 2019; Du et al., 2020). The fungus mostly affects immunocompromised patients, with mortality rates of up to 60% (Chen et al., 2020). According to the Centers for Disease Control and Prevention in the United States of America, echinocandins such as micafungin, caspofungin, and anidulafungin are recommended for the treatment of C. auris infections (Cândido et al., 2020).
Various other drugs, such as azoffluxin in combination with fluconazole, SS750, rocaglate, and colistin with echinocandins, have been shown to inhibit the growth of C. auris in different in vitro and in vivo experiments (Matsumoto et al., 2002; Ruiz-Gaitán et al., 2018; Bidaud et al., 2020; Iyer et al., 2020). Similarly, other drugs like sulfamethoxazole in combination with voriconazole, a low ionic solution formulation of caspofungin, and carvacrol (a polyphenol) have exhibited antifungal properties against C. auris (Eldesouky et al., 2018; Shaban et al., 2020; Sumiyoshi et al., 2020). Additionally, hydrogen peroxide vapor, chlorine, iodinated povidone, and chlorhexidine gluconate can kill C. auris, which can be beneficial in preventing nosocomial infections and transmissions (Abdolrasouli et al., 2017).
Numerous scientific studies have been conducted to discover novel drugs/phytochemicals to treat C. auris infections, but only a few have advanced to the clinical trial stage. Some antifungals that have progressed to clinical trials for C. auris include Ibrexafungerp, which inhibits 1,3-β-D-glucan synthesis, MGCD290, a fungal histone deacetylase (HDAC) inhibitor, VT-1129, which interferes with cytochrome P450 activity, and Fosmanogepix (APX001), which inhibits fungal glycosylphosphatidylinositol (GPI) anchor protein (Oliver et al., 2016; Ghannoum et al., 2020; Gintjee et al., 2020; Yu et al., 2020). However, the currently used antifungals such as polyenes, azoles, and echinocandins exert high selective pressure on the pathogen, leading to the development of drug resistance (Srinivasan et al., 2014).
Plants used in traditional and herbal medicine may be a key source of new antifungal compounds. Studies have shown that various herbs possess antimicrobial activity (Akhtar et al., 2017a, 2017b; Choudhury et al., 2017; Sarkar et al., 2021). Therefore, this study evaluated the antioxidant and antifungal properties of an extract from the ethnomedicinal plant Sarcochlamys pulcherrima. S. pulcherrima is an evergreen tree that grows in the forests of northeastern states of India, Thailand, Bhutan, Myanmar, and Indonesia (Mazumder et al., 2015), and is used by various tribes in India and Bangladesh to treat several ailments such as fever blisters, tongue ulcers, flatulence, boils, dysentery, diarrhea, digestion problem, and itching of the eyes (Mazumder et al., 2015). The antibacterial and antifungal properties of S. pulcherrima have been reported in previous studies (Mazumder et al., 2014; Ghosh et al., 2020).
In this study, prospective antifungals were investigated using the diffusion method, and the molecule was identified using high-performance thin-layer chromatography (HPTLC). The assay showed that gallic acid was one of the prominent molecules present in the S. pulcherrima leaves extract. We hypothesized that gallic acid was a potent antifungal and explored its efficacy against C. auris. Gallic acid has been found in a variety of plants and has been shown to possess antibacterial and antifungal activities (Li et al., 2017; El-Nagar et al., 2020; Simonetti et al., 2020). Additionally, the potential target of gallic acid in C. auris was determined using an in silico approach in this study.
The carbonic anhydrase (CA) enzyme can be a potential target for antifungal compounds. CA (EC number 4.2.1.1) is a metalloenzyme that efficiently catalyzes the interconversion of carbon dioxide (CO2) and bicarbonate (Elleuche and Pöggeler, 2010). It plays a crucial role in virulence in Cryptococcus spp. and C. albicans (Schlicker et al., 2009; Hall et al., 2010; Dostál et al., 2020; Fasciana et al., 2020), as well as phenotypic switching, sporulation, cAMP concentration increase, and survival in varying CO2 concentrations within the host (Buck and Levin, 2011; Huang, 2012; Jungbluth et al., 2012; Cummins et al., 2014). Due to its involvement in vital biological processes such as respiration, pH homeostasis, fungal growth, virulence, and CO2 transport, CA can be a crucial target for developing novel antifungal agents (Elleuche and Pöggeler, 2010; Supuran and Capasso, 2021). Previous studies have also suggested CA as a target for antifungal drugs (Schlicker et al., 2009; Supuran and Capasso, 2021). Therefore, in this study, we used a computational approach to assess whether gallic acid may function by binding with the CA of C. auris.
The study’s results suggest that gallic acid has the potential to impact the catalytic activity of the CA enzyme in C. auris and can be further investigated as a therapeutic agent for treating C. auris infections.
Materials and methods
Sample collection and preparation of extract
Leaves and barks from a single S. pulcherrima tree were collected from the New Halflong region of North Cachar Hills (Assam, India) in July 2018. The plant samples were washed thoroughly with water to remove any dust and impurities. The samples were air-dried in a hot air oven at 45–50°C until they were completely dry. The dried samples were then ground into a fine powder using a grinder, packed in airtight packets, and stored at 4°C for future use. The extract was obtained through Soxhlet-mediated extraction using two solvents, 90% methanol and ethyl acetate. To obtain the plant extract, 30 g of leaf powder was mixed with 100 ml of each solvent (Padmasree et al., 2017). The Soxhlet apparatus was run at ~65°C (methanol extraction) and ~77°C (ethyl acetate extraction) for 25 cycles (Padmasree et al., 2017). The crude solvent extract was then concentrated to 5 ml using a rotary evaporator at 40 rpm and 50°C temperature to remove excess solvent. The concentrate (5 ml) was further stored at 50°C to remove any remaining solvent until it reached a dry, paste-like consistency. The amount of the extract in paste form was measured and stored at 4°C until further use.
Determination of the antifungal activity of S. pulcherrima plant extract against C. auris
The C. auris strain National Culture Collection of Pathogenic Fungi (NCCPF) ID: 470097, which is sensitive to fluconazole and amphotericin B, was obtained from NCCPF, Post Graduate Institute of Medical Education and Research, Chandigarh, India. The antifungal activity of S. pulcherrima plant extract against C. auris was determined using the disc diffusion method. The yeast strains were grown in YPD broth overnight at 150 rpm shaking at 28°C. The overnight culture was then serially diluted to obtain 0.05 OD, and 100 μl of the diluted culture was spread over solidified Sabouraud dextrose agar (SDA). Filter discs containing different concentrations of the methanol and ethyl acetate plant extract were placed on the SDA plates. The plates were incubated for 48 h at 28°C. After the incubation period, the plates were observed, and the diameters of the zones of inhibition were measured. Fluconazole disc (Himedia, India, 10 μg), ethyl acetate, and methanol were used as controls. Each experiment was performed on three different petri plates with two biological replicates.
Determination of the antioxidant activity of S. pulcherrima plant extract
The in vitro antioxidant property of the S. pulcherrima leaf extract was determined through the 2,2-diphenyl-1-picrylhydrazyl (DPPH) free radical scavenging assay. For the assay, 0.135 mM DPPH was prepared in methanol (from a stock solution of 100 mM), and the S. pulcherrima leaf extract (previously extracted from methanol and ethyl acetate fractions) was prepared in dimethyl sulfoxide (DMSO) with concentrations ranging from 0 to 200 μg/ml. One milliliter of 0.135 mM DPPH and 1 ml of plant extract of varying concentrations were added to test tubes and incubated for 45 min in the dark at room temperature. All experiments were performed in triplicates, and ascorbic acid was used as a standard. Following the incubation, absorbance was measured at 517 nm, and the DPPH scavenging activity was calculated using the following formula:
whereAbsc is the average absorbance of methanol + DPPH and Abss is the average absorbance of sample (different concentrations of plant extract/ascorbic acid) + DPPH.
Determination of total phenolic content of S. pulcherrima plant extract
The phenolic contents in the S. pulcherrima leaf extract were estimated using the Folin-Ciocalteu reagent. Two hundred microliters of the leaf extract at different concentrations (160 and 320 μg/ml prepared in distilled water) were mixed with 2% sodium carbonate (2 ml) and 10% Folin-Coicalteu reagent (2.5 ml). The reaction mixture was then incubated at 45°C in a water bath for 20 min. After the incubation, the absorbance was measured at 765 nm. All experiments were performed in triplicate, and gallic acid (Sisco Research Laboratories, India) was used as a standard.
HPTLC detection of phenols in the plant extract
To identify specific phenolic compounds, such as gallic acid and quercetin, in the S. pulcherrima extract by Folin-Ciocalteu method, HPTLC analysis was performed at Herbal Health Research Consortium Private Limited in Amritsar, Punjab, India. A silica gel TLC plate coated with fluorescent indicator F254 measuring 5.0 × 10.0 cm (Merck, Germany) was used. The ethyl acetate extract of S. pulcherrima (2 μl) was applied using the CAMAG Linomat 5 semiautomatic sample application system in the form of bands (6 mm). After developing the TLC plates in a hexane-chloroform-acetic acid (4 : 5 : 1 ratio) solvent system, they were air-dried and scanned using a CAMAG TLC scanner at a wavelength of 270 nm with a Deuterium lamp in absorption mode. A gallic acid standard (Sisco Research Laboratories, India) of 1 mg/ml was used for the analysis.
Determination of in vitro antifungal activity of gallic acid against C. auris
The antifungal activity of gallic acid was evaluated against six clinical isolates of C. auris, which were obtained from NCCPF in Chandigarh, India. The identification numbers of these strains are 470,049, 470,055, 470,097, 470,098, 470,111, and 470,149. All C. auris strains used in the study were susceptible to fluconazole and amphotericin B, except of NCCPF 470149, which was resistant to fluconazole. Gallic acid was procured from Sisco Research Laboratories in India, and all experiments were conducted in triplicate.
The antifungal activity of gallic acid against C. auris strains was determined using the broth microdilution method based on EUCAST guidelines with modifications (Arendrup et al., 2012). Gallic acid stock solution was diluted in Sabouraud dextrose broth (SDB) to obtain concentrations ranging from 0.06 to 32 mg/ml. Next, 100 μl of the SDB with diluted gallic acid was added to respective wells in 96-well microtiter plates in triplicate, with an inoculum size of 0.5 × 104 CFU/ml. The plates were incubated for 24 h at 35°C without agitation, and the MIC (50%) was determined by measuring the absorbance at 530 nm, and calculated using the appropriate formula:
where Absc is the absorbance of well-containing media and culture.
The determination of the minimum fungicidal concentration (MFC) of gallic acid against C. auris followed the method described by Espinel-Ingroff et al. (2002). From the wells that showed complete inhibition (optically clear microtiter plate wells obtained after treating C. auris for the determination of MIC), 20 μl was spread plated on SDA plates and incubated at 35°C for 48 h. The MFC was determined as the lowest concentration that yielded less than three colonies (Espinel-Ingroff et al., 2002).
Structure prediction: modeling, validation, and superimposition of C. auris CA protein
The 3D structure of the C. auris CA protein has not yet been solved. The amino acid sequence of the C. auris CA protein (269 amino acids) was obtained from the Candida genome database (http://www.candidagenome.org/cgi-bin/locus.pl?locus=B9J08_000363), and the 3D structure of the C. auris CA protein was modeled using the I-TASSER web interface (Yang et al., 2015). I-TASSER employs a combination of different approaches, including threading, fragment assembly, ab initio loop modeling, and structural refinement (Yang et al., 2015). To identify the template protein structure, a BLAST search was conducted against the Protein Data Bank (PDB) database (Berman et al., 2000; McGinnis and Madden, 2004). The top hit obtained was the C. albicans CA protein with 79% sequence identity and 77% query coverage. Therefore, the crystal structure of C. albicans CA (PDB ID: 6GWU) was used as a primary template to model the C. auris CA protein structure. The best structure model was selected based on a confidence score (C-score).
To assess the quality of the modeled structure, we used the Structural Analysis and Verification Server (http://nihserver.mbi.ucla.edu/SAVES). Various structure assessment programs were used to analyze the backbone conformations and overall geometric compatibility of interacting residues in the modeled C. auris CA protein structure. The Ramachandran plot was used to estimate the phi/psi distributions of the protein backbone. The ERRAT program was used to calculate the nonbonded atomic interactions for the modeled proteins. The compatibility of the 3D atomic model of amino acid with its amino acid sequence (1D) was demonstrated by creating it through the verify-3D program.
To check the structural consistency, we superimposed the modeled structure with the template structure (6GWU) and calculated root mean square deviation (RMSD) using iPBA server (https://www.dsimb.inserm.fr/dsimb_tools/ipba/example.php). All molecular images were generated through UCSF Chimera software (Pettersen et al. 2004).
Molecular docking
The CA receptor and gallic acid ligand were subjected to molecular docking after energy minimization and refinement of both the modeled and crystal structures. The steepest descent (100 steps) and conjugate gradient (500 steps) algorithms were employed using the Swiss PDB viewer software platform (Guex and Peitsch, 1997). The Spatial Data File (SDF) structure of gallic acid was downloaded from the PubChem (https://pubchem.ncbi.nlm.nih.gov) database, with PubChem ID: CID 370. The downloaded SDF structure was converted to PDB format using the online SMILES translator and structure file generator (https://cactus.nci.nih.gov/translate/).
Firstly, the zinc metal-ion was docked in the modeled structure of C. auris CA protein using the Metal Ion-Binding (MIB) site prediction and docking server (http://140.128.63.8/MIB/). Gallic acid was then docked onto the Zn-coordinated C. auris CA using the Patchdock web server, with the criteria to generate a maximum of 10 docked poses (Schneidman-Duhovny et al., 2005). The active site amino acids in the C. auris CA modeled structure were marked based on the C. albicans CA structure (PDB ID: 6GWU). The docked protein–ligand complex was evaluated based on its atomic contact energy (ACE). The interacting residues of C. auris CA with the ligand were visualized using UCSF Chimera software (Pettersen et al., 2004).
Results
Determination of the antifungal activity of S. pulcherrima plant extract against C. auris
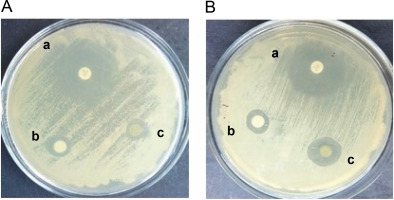
The antifungal activity of the methanol and ethyl acetate extracts of S. pulcherrima against C. auris (NCCPF ID: 470097) was determined using the disc diffusion method. The results showed that both extracts were able to inhibit the growth of C. auris, as demonstrated in Figure 1. The zones of inhibition for the methanol extract of S. pulcherrima against both C. albicans and C. auris strains were 11 mm. In contrast, the zones of inhibition for the ethyl acetate extract of S. pulcherrima against C. albicans and C. auris strains were 10 and 15 mm, respectively. This suggests that the ethyl acetate plant extract has better antifungal activity against C. auris than against C. albicans. The zones of inhibition for fluconazole against C. albicans and C. auris were 26 and 24 mm, respectively.
Antioxidant property of S. pulcherrima extract
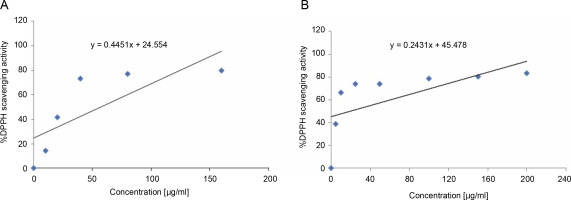
The S. pulcherrima leaves extract exhibited antioxidant activity by effectively scavenging the DPPH free radicals. The IC50 values of the extract and ascorbic acid were calculated from the graphs presented in Figure 2A and Figure 2B. The IC50 value of the S. pulcherrima leaves extract was found to be 57.16 μg/ml, while the IC50 value of ascorbic acid was 18.61 μg/ml.
Determination of total phenol content of S. pulcherrima plant extract
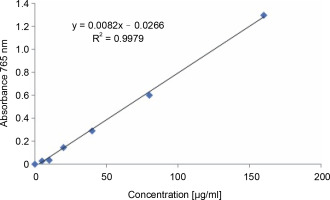
The total phenolic content of the methanol extract of S. pulcherrima leaves was determined by constructing a standard curve (Fig. 3) using gallic acid in the x-axis as the standard. The absorbance of the Folin-Ciocalteu reagent-gallic acid mixture at 765 nm was plotted in the yaxis against the concentration of gallic acid. The absorbance values of the S. pulcherrima extract for the determination of total phenolic content are provided in Table 1. The results showed that the phenolic content of the 160 μg/ml S. pulcherrima extract was equivalent to 40.31 μg/ml gallic acid, while the phenolic content of the 320 μg/ml S. pulcherrima extract was equivalent to 79.46 μg/ml gallic acid.
Detection of phenols in the S. pulcherrima plant extract
The presence of gallic acid and quercetin in the S. pulcherrima plant extract was analyzed by HPTLC. The gallic acid standard had a maximum Rf of 0.43 and a peak area of 16 416.6 (Supplementary Fig. 1A), while the S. pulcherrima plant extract showed a peak with a maximum Rf of 0.45 and a peak area of 1 431.3 (Supplementary Fig. 1B). This peak in the plant extract coincided with the peak of the gallic acid standard, indicating the presence of gallic acid in the S. pulcherrima plant extract. However, quercetin was not detected in the S. pulcherrima plant extract through HPTLC analysis.
Determination of in vitro antifungal activity of gallic acid against C. auris
The in vitro antifungal assay demonstrated that gallic acid was effective in inhibiting the growth of all six strains of C. auris used in the study. The minimum inhibitory concentration (MIC50) value of gallic acid, which is the lowest concentration that inhibited 50% of the growth of C. auris in comparison to untreated C. auris, was found to be in the range of 1.6–3.2 mg/ml. The MIC50 value of gallic acid (MIC50 GA) and MFC = lowest drug concentration that yields three or less colonies of gallic acid against the different C. auris strains are presented in Table 2.
Structure modeling and assessment of C. auris CA
The I-Tasser webserver was utilized to predict the 3D structure of the C. auris CA protein. The input for this analysis was the amino acid sequence of the C. auris CA protein in FASTA format. The C-score, a measure of the quality of predicted 3D models of proteins, for the modeled structure of the C. auris CA protein was −1.66 (Table 3A). The C-score values for protein models generated by I-Tasser usually range from −5 to 2. A higher C-score implies that the generated model is of higher quality (Yang et al. 2015). The modeled C. auris CA protein had a higher C-score, indicating that the predicted model was of good quality.
Table 3
Molecular modeling studies of C. auris CA protein
The quality of the predicted model of the C. auris CA protein was further assessed by generating a Ramachandran plot. Analysis of psi/phi angles showed that overall, 96% of residues in the modeled structure were present in the allowed regions of the Ramachandran map. The average ERRAT score of 93.10 for the modeled structure also confirmed the high accuracy of the predicted models. Additionally, the Verify3D software was used to assess the quality of the 3D model of the C. auris CA protein. This software assigns a structural class based on its location and environment (alpha, beta, loop, polar, non-polar, etc.) to determine the compatibility of the 3D model of proteins with its amino acid sequence (1D). The Verify3D software provided a 3D–1D score of 63.94% for the protein model, indicating that 63.94% of the amino acid residues of the C. auris CA protein are compatible with its 3D model (Table 3B).
Therefore, all three assessment methods, PRO CHECK, ERRAT, and Verify3D, confirmed the reliability of the predicted 3D structures. Superimposition of the modeled CA protein structure of C. auris and the experimentally determined CA structure of C. albicans showed an RMSD value (RMSD gives the average deviation between the corresponding atoms of two proteins and the lower RMSD value suggests the higher structural similarity between proteins) of 0.43 Å (Supplementary Fig. 2), further confirming the high accuracy of the modeled structure.
Molecular docking and evaluation of interacting residues
MIB web server predicted the binding residues i.e., for the positions 94 Cys, 149 His, and 152 Cys where the Zn2+ metal ion can bind in the C. auris CA protein. When the modeled structure of CA of C. auris was docked with Zn2+ metal ion, we found that all active site residues i.e., 94C, 149H, and 152C were interacting with it. The interacting residues are listed in (Table 4). The docking score and ACE which were obtained after docking the C. auris CA protein with gallic acid were 219 and −12.24 kcal/mol, respectively (Table 5). We also evaluated the docked complexes of both the CA proteins of C. auris and C. albicans with gallic acid using Chimera software (all within 5 Å from the ligand) and found significant changes in the binding residues and the neighboring residues (Table 6).
Table 4
Interacting residues and secondary structure pattern of C. auris and C. albicans CA protein within 5 Å from the Zn2+ metalion
Table 5
Docking studies of CA of C. auris and C. albicans yeast protein with gallic acid
| Protein | Ligand | Atomic contact energy [kcal/mol] | Docking score |
|---|---|---|---|
| CA of C. albicans | gallic acid | −71.85 | 1850 |
| CA of C. auris * | −12.24 | 219 |
Table 6
Interacting residues and secondary structure pattern of CA of C. auris and C. albicans yeast protein within 5 Å from the gallic acid
The MIB web server predicted the binding residues for the positions 94 Cys, 149 His, and 152 Cys, where the Zn2+ metal ion can bind in the C. auris CA protein. Docking the modeled structure of CA of C. auris with Zn2+ metal ion revealed that all active site residues (94C, 149H, and 152C) were interacting with it. The interacting residues are listed in (Table 4).
When the C. auris CA protein was docked with gallic acid, the docking score and ACE were 219 and −12.24 kcal/mol, respectively (Table 5). We evaluated the docked complexes of both the CA proteins of C. auris and C. albicans with gallic acid using Chimera software and found significant changes in the binding residues and the neighboring residues (Table 6). All residues within 5 Å from the ligand were analyzed.
When comparing the docking of gallic acid with the CA protein of C. albicans and C. auris, we found that the binding of gallic acid with C. albicans CA protein was energetically more favorable than with C. auris CA protein. The ACE for docking gallic acid with C. albicans CA protein was −71.85 kcal/mol, whereas the ACE for docking gallic acid with C. auris CA protein was −12.24 kcal/mol. The fact that the ACE was higher for C. auris CA protein suggests that the binding of gallic acid with C. albicans CA protein was more energetically favorable. Lower ACE indicates a more energetically favorable binding. However, in the docked complex of gallic acid with C. auris CA protein, all three desired active site residues (compared to active sites of C. albicans CA) did not interact with the ligand. Instead, only two active site residues (149H and 152C) were present in the docked complex. In contrast, when C. albicans CA was docked with gallic acid, we found interactions with all three desired active site residues. These observations indicate the presence of structural and mutational changes in the CA of C. auris.
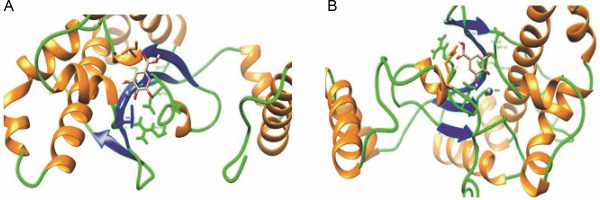
To verify this hypothesis, we analyzed the secondary structure distributions of all the interacting residues of both docked complexes with gallic acid (all within 5 Å from the ligand). We found a significant change in secondary structure in both docked structures (Fig. 4A and Fig. 4B). In the crystal structure of CA (PDB ID: 6GWU) of C. albicans, it has been reported that the active site residues (Ile 99, Gly 135, Phe 116, and Leu 121) create a narrow tunnel, which serves as the point of entry to the positively charged active site. The analysis of the modeled structure of C. auris CA protein showed that the Ile 117, Gly 153, Phe 134, and Leu 139 residues of C. auris CA protein also created a narrow tunnel. In C. albicans CA protein, there is a distinct salt bridge made up of residues present between arginine at position 80 and aspartic acid at position 133 (Arg80–Asp133) that contributes to the formation of the active site cavity (Dostál et al. 2018). The salt bridge regulates the tunnel’s shape and openness and contributes to the formation of the active site cavity in C. albicans CA protein (Dostál et al. 2018). A similar salt bridge was also present in the C. auris CA protein (Arg 98–Asp 151), but four positions had mutations (shown as double asterisk marks) (Supplementary Fig. 3). We infer that these mutations hinder the accessibility of gallic acid to the active site of C. auris CA protein, thereby decreasing the strength of binding.
Fig. 4
A) Interacting residues and secondary structure (Helix: Orange, Coil: Green, and Strand: Blue) pattern of C. albicans CA protein within 5 Å from the ligand (gallic acid); B) Interacting residues and secondary structure (Helix: Orange, Coil: Green, and Strand: Blue) pattern of C. auris CA protein within 5 Å from the ligand (gallic acid)
Discussion
C. auris infections present a severe global public health threat due to their high mortality rate, rapid transmission, and multidrug resistance (Chowdhary et al., 2017; Giarratano et al. 2018). Since its first identification in 2009, C. auris infections have caused hospital outbreaks in over thirty countries on six continents (Rhodes and Fisher, 2019). Besides multidrug-resistant strains, different C. auris strains exhibit pan-resistance, which is resistance to all three major classes of antifungal drugs: azole, echinocandins, and polyene (O’Brien et al., 2020; Ostrowsky et al., 2020). Furthermore, the side effects and low bioavailability of existing antifungals (Thammahong et al., 2017; Akhtar et al., 2021) can further hinder the treatment of C. auris infections. Therefore, it is necessary to continue searching for new, effective, and safe antifungal drugs. Plants can be an essential source of novel drugs for treating various ailments. Humans have been using plants and plant-based compounds as part of various traditional medicinal practices, such as traditional Chinese medicine, Unani medicine, Iranian traditional medicine, Siddha medicine, Kampo medicine, and Ayurveda (Akhtar et al., 2021). Plants are rich in a plethora of metabolites belonging to different classes such as alkaloids, flavonoids, phenolic acids, and saponins (Akhtar et al., 2021). More than 50 000 secondary metabolites have been discovered in various plants, and modern medicine relies on these metabolites (Teoh, 2016). Several metabolites found in plants, such as saponins, flavonoids, polyphenols, terpenoids, tannins, alkaloids, and coumarins, have been reported to possess antifungal properties (Castillo et al., 2012; Mishra et al., 2020; Akhtar et al., 2021). In this study, we evaluated the antifungal property of S. pulcherrima leaves’ extract against C. auris, identified the active compound responsible for the antifungal property, and determined the potential mode of action of the active compound by in silico approach.
Previous studies have reported the antioxidant and antifungal properties of S. pulcherrima methanolic extract against C. albicans (Paul et al., 2010; Mazumder et al., 2012; Mazumder et al., 2014). However, the modes of action and efficacy of the extract against multidrug Candida spp. were not explored. In our study, both the methanol and ethyl acetate extract exhibited antioxidant and antifungal properties against C. auris and C. albicans. After determining the antifungal and antioxidant properties of the plant extract, we conducted experiments to identify the potential active metabolite in the extract.
Previously, aerial parts of S. pulcherrima were found to contain two triterpenoids, tormentic acid, and 23-hydroxycorosolic acid (Ghosh et al., 2020), which inhibited biofilm formation by Pseudomonas aeruginosa (Ghosh et al., 2020). Given the presence of phenolic compounds with antioxidant properties in the plant extract, we performed an HPTLC experiment to identify potential antioxidant compounds, such as quercetin and gallic acid. The HPTLC analysis confirmed the presence of gallic acid in the extract. Gallic acid is a phenolic compound naturally found in various herbal plants and fruits, such as Camellia sinensis, Oenothera biennis, Vitis vinifera, Terminalia bellerica, and Terminalia chebula (Karamać et al., 2006; Genwali et al., 2013; Singh et al., 2019). Gallic acid has been reported in in vitro and in vivo studies to possess various therapeutic potentials, including antimicrobial, antioxidant, anti-inflammatory, anticancer (Choi et al., 2009; Giftson et al., 2010a; Shao et al., 2015; Kahkeshani et al., 2019; Yang et al., 2020), chondroprotective, hepatoprotective, neuroprotective, and antidiabetic properties (Kroes et al., 1992; Giftson et al., 2010b; Patel and Goyal, 2011; Huang et al., 2012; Nabavi et al., 2012; Gandhi et al., 2014; Bayramoglu et al., 2015; Wen et al., 2015).
An FDA-unapproved homeopathic drug manufactured by BioActive Nutritional, Florida, USA, with gallic acid as the active ingredient, has been marketed for temporary relief from itching, back pain, joint pain, nasal congestion, fatigue, allergic rhinitis, and dyspepsia (DailyMed, 2013). However, the safety and efficacy of this homeopathic drug have not been evaluated by the FDA (DailyMed, 2013) in vitro studies have reported the antifungal activity of gallic acid against different human fungal pathogens, such as C. albicans, Candida krusei, Candida glabrata, Candida tropicalis, Microsporum canis, and Trichophyton species (Li et al., 2017; Carvalho et al., 2018). In a study conducted by Li et al. (2017), the MIC of gallic acid against C. glabrata, C. albicans, and C. troplicalis was reported to be between 12 and 100 μg/ml. Similarly, another study reported that the MIC of the gallic acid against C. albicans, C. krusei, C. tropicalis, and C. glabrata as well as dermatophytes such as M. canis and Trichophyton species, ranged between 0.25 and 1 mg/ml (Carvalho et al., 2018). In our study, gallic acid exhibited antifungal activity against C. auris, with a MIC ranging from 1.6 to 3.2 mg/ml, with four strains having a MIC of 1.6 mg/ml and the remaining two C. auris strains having a MIC of 3.2 mg/ml. This is consistent with previous studies that reported the antifungal activity of gallic acid against different human fungal pathogens such as C. albicans, C. glabrata, C. krusei, and C. tropicalis, as well as dermatophytes such as M. canis and Trichophyton species (Li et al., 2017; Carvalho et al., 2018). Furthermore, the administration of gallic acid to immunocompromised mice models with systemic C. albicans infection has been reported to reduce mortality rate, inflammation, and fungal load in liver tissues of mice, corroborating the hypothesis that gallic acid is one of the potential active compounds in S. pulcherrima responsible for its antifungal property (Li et al., 2017).
We conducted in silico experiments to investigate the potential mode of action of gallic acid. Prior studies have reported that the inhibition of ergosterol biosynthesis and reduction in CYP51 enzyme activity are the mechanisms behind the antifungal activity of gallic acid (Li et al., 2017; Carvalho et al., 2018). In our study, we explored the CA protein of C. auris as a potential target of gallic acid, which is responsible for virulence and various other important biological functions in different fungi (Schlicker et al., 2009; Elleuche and Pöggeler, 2010; Supuran and Capasso, 2021). Targeting virulence proteins could minimize the adverse effects on host cells and limit the emergence of drug resistance in microbes by reducing the selective pressure (Marra, 2004; Heras et al., 2015). Therefore, CA can be a promising target for discovering novel antifungal drugs.
In a previous study, the repurposing of FDA-approved CA inhibitors such as ethoxzolamide, acetazolamide, and methazolamide, which were used to treat glaucoma or duodenal ulcers, demonstrated antibacterial activity against Vancomycin-resistant Enterococcus faecalis and Enterococcus faecium (Kaur et al., 2020). The molecular docking and molecular dynamics study indicated that the CA of E. faecium could be the intracellular target of ethoxzolamide, acetazolamide, and methazolamide (Kaur et al., 2020).
Our study suggests that gallic acid can bind with the active site residues of CA proteins from C. auris and C. albicans. The majority of the active site residues are hydrophobic (Ile 99, Gly 135, Phe 116, and Leu 121), forming a narrow tunnel that serves as the point of entry to the positively charged active site in a C. albicans crystal structure (6GWU). In the simulated structure of C. auris, we found that these residues (Ile 117, Gly 153, Phe 134, and Leu 139) were present in the protein as well, forming a small tunnel. The distinct salt bridges of 54 residues (Arg80–Asp133) that contribute to the formation of the active site cavity in C. albicans and regulate the shape and openness of the tunnel were also present in C. auris (Arg98–Asp151). However, at four positions, mutations were found in C. auris CA protein (Supplementary Fig. 3). The residues Asp 83, Val 95, Ile 118, and Lys 124 of C. albicans CA protein were mutated to Glu 101, Thr 113, Val 136, and Arg 142, respectively, in C. auris CA protein. We believe that these changes make gallic acid less accessible to the active site, lowering the binding strength. Future studies can focus on developing derivatives of gallic acid that can target the CA of C. auris more efficiently, leading to the development of novel antifungal drugs with new modes of action.
Conclusion
The antifungal analysis conducted in this study suggests that S. pulcherrima, also known as Mesaki, could serve as a potential source of novel antifungal drugs, with gallic acid being one of the active molecules responsible for its antifungal properties. The MIC of gallic acid against C. auris ranged from 1.6 to 3.2 mg/ml. Our in silico experiments revealed that gallic acid could bind to the CA protein, which is essential for fungal virulence in C. auris and C. albicans. Moreover, for the first time, we predicted the structure of the CA protein from C. auris. Our research showed that gallic acid from Mesaki tree leaves could potentially combat fluconazole-resistant C. auris clinical isolates. However, further in vivo and clinical studies are required to confirm gallic acid’s antifungal properties. In the future, derivatives of gallic acid with enhanced antifungal potential targeting the CA protein could be developed, leading to the emergence of a new class of antifungal medications that target the CA protein.