Introduction
Worldwide, more than 90% of food needs are met by 12 crop varieties and 14 animal species (Masciarelli et al., 2014; Ashraf et al., 2019; Bhardwaj and Kumar, 2020). More than half of the global food demand is fulfilled by wheat, rice, and maize (Macauley, 2015). Climate change and abiotic stresses generated by soil salinity are the principal issues encountered by the modern agriculture sector, adversely affecting soil fertility and productivity (Sahab et al., 2021; Zuo et al., 2021). A previous study has documented that climate change may increase the occurrences of heat stress, which decreases groundwater levels and increases salinity levels in the soil (Singh et al., 2015). An increase in ground-water evaporation and a reduction in rainfall increase salinization and alkalization worldwide (Eswar et al., 2021; Wang et al., 2021). Subsequently, an increase in subsoil drying increases the accumulation of salts in groundwater. This effect turns fertile fields into arid lands and reduces crop growth, yield, flowering, and pollination (Mutuku et al., 2020; Zuo et al., 2021; Wang etal., 2021). Soil salinity affects the plant growth at later stages, as it interferes with root turgor, leading to a reduction in water absorption; a decrease in the plant water column that progresses through dehydration and osmotic stress; inhibition of the metabolic machinery; disturbance in the transpiration system; and most importantly, interference with parameters of photosynthesis (Kaushal and Wani, 2016).
Plants share a significant proportion of their resources with various microorganisms in their natural habitat (Yadav et al., 2020). Plants and microbes such as bacteria may form beneficial mutual partnerships in this nutrient-rich environment (Ashraf et al., 2019). These bacteria are involved in a variety of metabolic pathways that assist plants in coping with stress. Plant-growth-promoting bacteria (PGPB) are found in various physical and chemical environments and are associated with various plant species in free-living or symbiotic relationships (Ahmed, 2018; Ashraf et al., 2019). Plant growth promoting rhizobacteria (PGPRs) (Glick et al., 1998) are a subclass of PGPB and one of the most thoroughly researched plant-associated microbial communities found on root surfaces and at deeply adhering soil interfaces. To improve plant growth and disease management, free-living and symbiotic bacteria use various pathways, i.e., nitrogen fixation, siderophore production, phosphate solubilization, improvement in nutrient uptake, phytohormone production, and antifungal activity (Almaghrabi et al., 2013; Masciarelli et al., 2014; Zhou et al., 2017; Etesami and Maheshwari, 2018). Many studies (Masciarelli et al., 2014; Zhou et al., 2017; Karnwal, 2017) have reported that PGPRs are efficient during sudden environmental changes, especially abiotic stresses. This stress-tolerant characteristic could be crucial in synthesizing biofertilizers that are used as PGPB. Such microorganisms help reduce salinity stress and restore salinity-sensitive land for agricultural purposes (Karnwal and Dohroo, 2018; Kumar and Verma, 2018). PGPRs are also involved in plant development by decreasing organic matter, detoxifying, and reducing plant infections (Etesami and Maheshwari, 2018; Ashraf et al., 2019; Sahab et al., 2021). Flavobacterium, Azotobacter, Bacillus, Pseudomonas, Enterobacter, Mesorhizobium, Mycobacterium, Burkholderia, Erwinia, and Rhizobium are the most prevalent PGPB genera found in soil (Karnwal, 2017; Bhardwaj and Kumar, 2020).
Crops adapt to natural evolution and revolutionize themselves viably using multiple approaches such as nutrient absorption, maintaining ionic equilibrium (Kumar and Verma, 2018), physiological reactions, antioxidant production (Karnwal, 2019), and proline accumulation to counteract salt toxicity. More than 36% of the global population consumes wheat as the primary staple food. Wheat comprises about 55% of the carbohydrates consumed globally and 20% of the calories in seeds (Macauley, 2015). Wheat production is adversely affected by saline conditions, as they lead to decreased seed germination, abnormal reproduction, reduced plant growth and enzyme activity, poor photosynthesis, phytohormone imbalance, and oxidative stresses (Ramadoss et al., 2013; Singh et al., 2015; Ashraf et al., 2019). Similarly, maize is cultivated in a diverse range of soil types and climates and is highly susceptible to salt stress (Macauley, 2015; Hussain et al., 2019); hence, soil salinity presents a significant challenge to the productivity of both crops globally.
The species of Bougainvillea grow and flourish in a saline environment (Islam et al., 2016). They are extremely drought tolerant, salt tolerant, and wind resistant. Bougainvillea glabra thrives in well-drained acidic soils with a pH of 5.5–6.0 (Abarca-Vargas and Petricevich, 2018; Karnwal, 2021). In contrast, wheat and maize usually grow under nonsaline/nonstress conditions, so using these cereal crops to isolate salt-tolerant bacteria may not be feasible. Islam et al. (2016) reported that Bougainvillea species, associated with several salt-tolerant plant growth promotion (PGP) strains in the root-associated soil, can be used for plant growth improvement under saline conditions (Abarca-Vargas and Petricevich, 2018; Karnwal, 2021). The present study aimed to culture and screen salinity-tolerant PGPRs of B. glabra rhizosphere and examine their growth-promoting activity with wheat (Triticum aestivum ) HD-2687 and maize (Zea mays ) PSCL-4642 cultivars under saline conditions at the seedling stage.
Materials and methods
Isolation and purification of salt-tolerant rhizobacteria from B. glabra
Rhizobacterial isolates were cultured from B. glabra rhizosphere (latitude 30.952802, longitude 76.776914) (Karnwal, 2017). On 4% salt (NaCl)-amended nutrient agar medium (NAM), 0.1 ml of serially diluted rhizospheric soil sample suspension was spread. The plates were incubated at 28 ± 1°C for 72 h, and their macroscopic and microscopic characteristics were examined. The pure cultures (overnight-grown bacterial cultures) were subsequently preserved with 30% glycerol in a deep freezer (Meiling Inc.) at !80EC until further use.
Screening of salt-tolerant bacterial isolates at different salt levels
Isolated bacterial species were examined for their salt tolerance potential on the NAM amended with different sodium chloride concentrations (5, 7, 8.5, 10, and 12%). Bacterial isolates that showed tolerance to 4% NaCl were selected and further examined for in vitro plant-growth-promoting activities (Karnwal, 2019). For cluster analysis, bacterial growth at different salt concentrations was scored with binary numbers (1 for growth and 0 for no growth). The recorded binary data were analyzed using the neighbor-joining algorithm for dendrogram preparation using the PAST (PAleontological STatistics) software, version 3.22. Of the 24 isolates, five (BoGl109, BoGl113, BoGl120, BoGl123, and BoGl124) grew above 4% salt stress, and their morphological, biochemical, and PGP characteristics were further studied.
Biochemical characterization
The five bacterial isolates selected (Karnwal, 2021) (BoGl109, BoGl113, BoGl120, BoGl123, and BoGl124) were subjected to microscopic (gram staining) and biochemical (casein hydrolysis, catalase production, citrate utilization, gelatine liquefaction, hydrogen sulfide (H2S) production, hydrogen cyanide (HCN) production, indole production, lipolysis activity, starch hydrolysis, oxidativefermentative reaction, and urease production) analysis in accordance with Bergey’s Manual of Determinative Bacteriology (Holt et al., 1994).
Screening for potential plant-growth-promoting isolates
Screening for the synthesis of indole acetic acid (IAA)
Bacterial isolates for IAA phytohormone synthesis were examined in the Dworkin and Foster (DF) broth supplemented with L-tryptophan (0.1%) following the Salkowski method (Dworkin and Foster, 1958; Schwyn and Neilands, 1987; Karnwal, 2017). Of the bacterial culture, 100 μl was transferred individually and incubated in DF broth at 28 ± 1°C for 72 h. The culture broth was then centrifuged at 11 963 g, and the supernatant was collected. Of the culture supernatant, 1 ml was mixed with 2 ml of Salkowski reagent and left for 30 min at 28± 1°C in a dark environment. IAA-like auxins thus produced were measured at 530 nm using a UV-Vis Spectrophotometer (GENESYS 40/50 Vis/UV-Vis Spectrophotometer). The amount of IAA was measured by a standard curve prepared using pure IAA (Hi-media) solutions ranging from 0 to 250 μg/ml of pure IAA (Almaghrabi et al., 2013).
Phosphate solubilization
Phosphate solubilization of each of the bacterial isolates was evaluated using the spot inoculation technique on Pikovskaya’s agar and reported as the solubilization index (SI) (Karnwal, 2017). Bacterial colonies producing a clear phosphate-solubilizing zone were considered positive for phosphate solubilization and quantified using the following equation (Premono et al., 1996):
Screening for siderophore production
The siderophore production potential of the five bacterial isolates was determined using the chrome-azurol-S agar (CAS) medium as described by Schwyn and Neilands (1987). CAS agar plates with bacterial isolates were incubated at 28 ± 1°C for 72 h. Changes in the CAS medium around bacterial growth from blue to yellow or orange were recorded as positive for siderophore production.
HCN production
All five bacterial isolates were subjected to the Bakker and Schippers (1987) assay method for HCN synthesis. L-Glycine (4.4 g/l)-supplemented modified NAM was used for HCN production. The inner surface of the upper lid of Petri plates was covered with filter paper (Whatman No. 1) dipped in 0.5% picric acid in 1% Na2CO3. For HCN synthesis, bacterial colonies were transferred to modified NAM plates along with the uninoculated control. Petri plates were sealed with Parafilm and incubated at 28± 1°C until a light, moderate, or dark brown color developed, which confirmed HCN synthesis.
Of the five tested isolates, BoGl123 showed the highest PGP activity and maximum salt tolerance capabilities. Thus, it was selected for root colonization, seed inoculation, and molecular identification studies.
Colonization ability
Root colonization of BoGl123 around wheat and maize seeds was tested following the method of Chauhan et al. (2016). Crop seeds were procured from the Indian agricultural research institute (IARI), Delhi, India (Karnwal, 2019). They were surface-sterilized using 3% sodium hypochlorite (NaOCl). They were immersed in NaOCl for 6 min and gently rinsed thrice with sterile distilled water (DW) to remove traces of NaOCl. The surface-sterilized seeds were then placed under laminar airflow for air drying, inoculated in presterilized agar plates, and incubated at 28 ± 1°C for 24 h to test the efficacy of the sterilizing agent. Finally, the colonization ability of BoGl123 was evaluated as described by Karnwal (2021).
Seed inoculation
The crop seeds were surface-sterilized using 3% NaOCl. They were immersed for 5 min in NaOCl and then gently rinsed thrice with sterile DW to remove traces of the sterilizing agent. To test the sterilizing agent’s efficacy, the surface-sterilized dried seeds were inoculated in presterilized NAM plates and incubated at 28± 1°C for 24 h. After 24 h, no microbial growth was observed on seed-inoculated plates, and the seeds were immersed in bacterial suspension (cell density 108 CFU/ml at 600 nm) for 2 h at 28 ± 1°C and air-dried under aseptic conditions (Karnwal, 2019).
Bacteria seed inoculation under salinity conditions (50, 100, 150, and 200 mM)
Different NaCl concentrations were used to examine the effect of BoGl123 on the germination of seedlings, as reported by Karnwal (2021). To preserve salinity throughout the study, the surface-sterilized seeds were placed on Petri dishes containing presterilized moist filter paper (Whatman-no. 1) with 5 ml sterile NaCl (50, 100, 150, and 200 mM) solution (10 seeds on each plate). Sterile DW was used as the control with equal salinity conditions (50, 100, 150, and 200 mM). Kader’s equation was used to calculate the germination percentage (%):
Molecular identification
The bacterial culture was outsourced for identification and 16S RNA sequencing to Chromous Biotech, Bangalore, India. Phylogenetic analysis of the 16S rRNA sequence of BoGl123 was conducted as described by Karnwal (2019). Genomic DNA Mini Spin Kit (Chromous Biotech, Bangalore, India) was used to isolate BoGl123 genomic DNA. Of the bacterial pellet, 100 mg was gently mixed with 1× suspension buffer (750 μl) and 5 μl RNase A, subjected to random stirring, and incubated for 10 min at 65EC. The mixture was intermittently mixed with 1 ml lysis buffer and kept at 65EC for 15 min. The mixture was centrifuged at 13 000 g in a 2 ml sterilized vial, and the supernatant was collected. Then, 600 μl of the supernatant was recentrifuged at 25EC for 1 min at 13 000 g onto the spin column. The content in the collection tube was removed, and the column was filled with 500 μl 1× wash buffer and centrifuged at 13 000 g at 25EC for 3 min. The spin column was transferred in a presterilized 1.5-ml collection tube to which a warm (65EC) elution buffer (50 μl) was added. The collection tube was centrifuged at 13 000 g for 1 min at 25EC. Agarose gel electrophoresis (1%) was used for the collection and resolution of the eluted DNA sample.
Polymerase chain reaction (PCR) amplification was carried out in a 100-μl PCR mixture containing 1μl DNA, 400 ng dNTPs (2.5 mM each), 400 ng of each primer, 3 U of Taq DNA polymerase, and 10 μl 10 × Taq DNA polymerase assay buffer. The universal bacterial forward: 5´–AGHGTBTGHTCMTGNCTCAS–3´ and reverse primers: 5´–TRCGGYTMCCTTGTWHCGACTH–3´ were used for PCR amplification. The initial denaturation period of each PCR cycle was 5 min at 95EC and a 30-s denaturation stage, followed by a 35-step amplification process at 94EC, a 30-s annealing stage at 50EC, and a 30-s extension stage at 72EC. PCR amplifications were completed with 7-min elongation at 72EC. Agarose gel electrophoresis (1% (w/v)) (200 V) was used for the collection and resolution of PCR products. After purification and amplification, the BigDye Terminator v3.1 Cycle Sequencing Kit was used to sequence the PCR products with an ABI 3500 Genetic Analyzer. The 16S rRNA sequence of the isolate was then subjected to an nBLAST (Nucleotide Basic Local Alignment Search Tool) search against the NCBI (National Center for Biotechnology Information) database for nucleotide sequence homology for the 16S region for bacteria. Multiple sequence alignment for sequences collected from the nBLAST search was carried out using the MUSCLE (MUltiple Sequence Comparison by Log-Expectation) algorithm. Then, the sequence of BoGl123 was subjected to the evaluation of the evolutionary distance of the stress-tolerant strain with the collected sequences and phylogenetic analysis using the MEGA X software.
Statistical analysis
All experiments were performed in triplicate using a completely randomized block design. Numerical values obtained from seed germination and growth promotion trials were subjected to analysis of variance. Fisher’s significant difference test was used to compare the treatments’ means using P values of 0.05.
Results and discussion
Plants have to tolerate multiple biotic and abiotic stresses under real-world conditions (Singh et al., 2015). Abiotic stress is a nonbiological environmental factor that substantially influences crop productivity in the agricultural industry. It affects factors such as nutrients, salt, water, temperature, and pH (Kumar and Verma, 2015). In the present work, 24 bacterial isolates from B. glabra rhizosphere (labeled from BoGl101 to BoGl124) were examined for salt tolerance in a 4% NaCl-supplemented nutritional agar medium.
Screening of bacterial isolates with salt stress tolerance
Microbes follow a range of strategies to thrive in stressful environments (Balseiro-Romero et al., 2015), namely, through the production of a large volume of exopolysaccharides (Torres et al., 2019). In the present study, 24 bacterial isolates were grown with an NaCl concentration of more than 4% on Petri plates, which was tolerated by five isolates (BoGl109, BoGl113, BoGl120, BoGl123, and BoGl124). BoGl123 showed growth up to a maximum of 10% salt concentration, whereas the other four isolates showed growth only up to 8.5% salt level. Recently, good abiotic stress tolerance potential was reported for many bacterial species belonging to the genera Achromobacter, Bacillus, Burkholderia, Methylobacterium, Pseudomonas, Rhizobium, and Variovorax (Passari et al., 2016; Armada et al., 2016; Karnwal, 2019). These microbes could help overcome environmental factors in agriculture by resolving soil salinity issues during crop production.
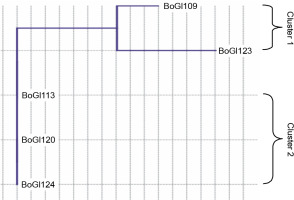
A dendrogram for salinity stress tolerance at the different NaCl levels for all five isolates was prepared using the neighbor-joining method (similarity index Euclidean). Two clusters were distinguished: cluster I included BoGl109 and BoGl123 and cluster II included BoGl113, BoGl120, and BoGl124, as shown in Figure 1. Cluster analysis demonstrated that BoGl109 and BoGl123 better tolerated salinity than BoGl113, BoGl120, and BoGl124; however, only BoGl123 was able to tolerate a maximum of 10% NaCl, wherein slow growth was observed (required longer incubation time), whereas, at 12% concentration, no growth was observed.
Plant growth promotion
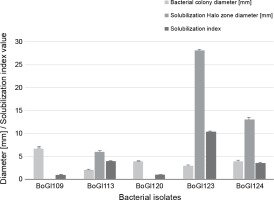
The salt stress tolerance experiments were conducted to examine the IAA production, inorganic phosphate solubilization, siderophore production, and HCN synthesis of the five bacterial isolates. All living organisms need phosphorus for development and survival (Saleem et al., 2019). Plants need phosphorus at modest levels, and its decreased bioavailability might lead to decreased growth and productivity of plants (Karnwal, 2017). The optimal amount of phosphorus needed for plant development varies from 25 to 30 μmol/l, but it usually ranges from 1 to 1.7 μmol/l in most of the soil types (Rahmoune et al., 2017). Several studies (Almaghrabi et al., 2013; Kumar and Verma, 2018; Torres, 2019) have demonstrated that plants use soil microbes to convert inorganic phosphates into accessible forms. The results of the present study revealed BoGl113, BoGl123, and BoGl124 as positive isolates for phosphate solubilization, with the formation of a visible halozone surrounding the bacterial growth with a diameter of 6–28 mm and an SI of 3.60–10.33, indicating positive results (Fig. 2).
IAA is a plant growth hormone associated with root development and proliferation (Karnwal, 2009; Karnwal and Dohroo, 2018). In the present study, three isolates showed promising IAA production, as shown in Table 1. The maximum IAA production was observed in BoGl123 (8.23 μg/ml), followed by BoGl113 (1.6 μg/ml) and BoGl120 (6.23 μg/ml).
Table 1
Results of plant-growth-promoting properties of rhizobacterial isolates
Karnwal (2009) discussed the significance of rhizospheric stress-tolerant competent bacteria with a diverse set of PGP characteristics that aid plants in dealing with salt stress. One of the essential traits of PGPRs is their ability to synthesize iron-chelating siderophores (Chauhan et al., 2016). Siderophores promote plant development by either increasing or decreasing the iron supply for phytopathogens (Schwyn and Neilands, 1987; Etesami and Beattie, 2018). In the present study, four isolates tested positive for HCN production (BoGl109, BoGl113, BoGl120, and BoGl123), whereas all five tested positive for siderophore production (Table 1). Sadeghi et al. (2012) evaluated the application of Streptomyces sp. in wheat cultivation under a saline soil environment and found that Streptomyces sp. can produce auxins and siderophores and improve seed germination, rhizome length, dry weight, shoot length, and dry weight compared with uninoculated control treatments.
Of the five isolates, BoGl123 showed the highest salt tolerance (up to 10% NaCl level) and plant-growth-promoting features. As a result, the BoGl123 strain was selected for further research, including root colonization by bacterial isolates, bacterial seed inoculation and seed germination under salinity conditions, and 16S-rRNA-based molecular identification of the BoGl123 strain.
Root colonization
BoGl123 was found to harbor the radicles of both experimental crop seedlings. It generated a thick hazy zone surrounding the roots and adjacent to the roots. The presence of the hazy zone is an indicator of bacterial root colonization.
Seed inoculation and germination under salinity conditions
Under real-world conditions, plants are continuously subjected to various abiotic stresses; in particular, salinity is one of the most severe factors responsible for reduced development and yield in the agricultural sector. Inoculation of PGPRs has been reported as a successful method (Etesami and Maheshwari, 2018) to improve plant productivity under saline stress conditions. Therefore, it is critical to analyze the relationships between the host plant and microorganisms. Previous studies have reported that pretreatment of seeds with different PGPRs promotes seed germination and seedling growth (Rahmoune et al., 2017; Bakhshandeh et al., 2020). In addition, enhanced dried biomass and rhizobacterial communities are found to boost root and soil microbial respiration in a saline environment, which in turn affects soil atmosphere composition. The findings of the effect of salinity stress and inoculated bacteria on seed germination of wheat and maize at different concentrations of NaCl are presented in Table 2 and Table 3. A positive influence of the bacterial inoculant on the percentage of germinating seeds was observed compared with the control. A previous study reported that uninoculated seedlings were more vulnerable to stress impact due to the lack of salinity-tolerant bacterial communities (Sen et al., 2018).
Table 2
Efficacy of bacterial treatment on germination of seeds at different NaCl concentrations
Table 3
Efficacy of bacterial treatment on radicle length (mm) at different NaCl concentrations
Moreover, the application of salinity-tolerant Pseudomonas spp. may have increased the root exudation, which increased the root colonization of the inoculated bacterial community and enhanced the plant growth. Alori et al. (2020) reported that two Pseudomonas strains (Pseudomonas kilonensis F113 and Pseudomonas protegens CHA0) substantially improved the growth of maize in pot experiments with an enhanced shoot, leaf, and root length. In the present study, BoGl123 improved the percentage of seed germination in both crops compared with the control. Inoculation with BoGl123 resulted in 100% (50 mM NaCl conc.), 98% (100 mM NaCl conc.), and 64% (150 mM NaCl conc.) seed germination in wheat and 100% (50 mM NaCl conc.) and 86% (100 mM NaCl conc.) seed germination in maize. These findings support the beneficial impact of bacterial inoculation on seeds (Table 2). Egamberdieva and Lugtenberg (2014) and Habib et al. (2016) reported that Pseudomonas strains (Pseudomonas aurantiaca TSAU22, Pseudomonas extremorientalis TSAU6 and P. extremorientalis TSAU20) and Enterobacter sp. UPMR18 improved wheat plant growth compared with control plants and alleviated salinity (100 mM NaCl)-induced dormancy of wheat seeds. Singh and Jha (2016) demonstrated the positive effect of inoculation of Serratia sp. Sl-12, a halophilic bacterium, on wheat plants (T. aestivum ), which resulted in higher salt resistance and shoot biomass. Upadhyay and Singh (2015) reported that inoculation of Bacillus aquimaris strains with wheat seeds in saline environments resulted in an increased total soluble sugar content and a higher shoot biomass production. Rajput et al. (2013) demonstrated improvement in wheat development and productivity when inoculated with the alkaliphilic bacterium Planococcus rifietoensis. Sharma et al. (2016) extracted five salt-tolerant bacterial genera from roots of Arthrocnemum indicum, with a salt tolerance potential of up to 8% NaCl concentration, and determined that they belonged to the bacterial genera Agrobacterium, Klebsiella, Ochrobactrum, and Pseudomonas. They reported that inoculating these salt-tolerant bacterial genera in saline soil improved the growth and yield of peanut.
BoGl123 treatment considerably enhanced the radicle length of wheat seedlings under different salinity stresses, as shown in Table 3. It enhanced the radicle length at 50, 100, and 150 mM salinity levels by 34, 21.2 and 1.2 mm, respectively. In addition, in maize, BoGl123 improved the radicle length by 26.8 and 14.0 mm at 50 and 100 mM salinity levels. The results of the present study are in accordance with those of Ramadoss et al.’s (2013) study. Ramadoss et al. (2013) investigated the influence of five halotolerant PGPRs (Bacillus and Halobacillus genera) on T. aestivum L. seedlings. They reported the effect of bacterial inoculation on the mitigation of salinity stress in T. aestivum L. seedlings at 80, 160, and 320 mM salinity levels. The inoculation resulted in a 71.7% improvement in the root length compared with uninoculated positive controls. Furthermore, tomato (Solanum lycopersicum) seedlings inoculated with Pseudomonas putida UW4 showed significantly increased shoot and root development after 6 weeks under saline conditions (90 mM NaCl conc.) (Yan et al., 2014). Zhang et al. (2018) documented the heterogeneity of salt-tolerant bacteria extracted from the rice rhizo-spheric soil in Taoyuan, China. They reported the presence of 162 salt-tolerant bacterial strains able to tolerate 150 g/l salt concentration. These salt-tolerant isolates improved the development and productivity of paddy in a salt-stress environment. Kearl et al. (2019) extracted 41 salt-tolerant isolates from Allenrolfea occidentalis, Salicornia rubra, and Sarcocornia utahensis halophyte plants. These bacterial isolates showed growth in 4 M NaCl-amended nutrient media. Halomonas, Bacillus, and Kushneria were the major salt-tolerant bacterial genera reported in their study, of which Halomonas and Bacillus isolates stimulated plant growth of alfalfa seedlings when applied under 1% NaCl saline conditions.
BoGl123 enhanced the plumule length of the wheat and maize germinated seedlings by 38.2 and 30.2 mm at 50 mM NaCl, respectively (Table 4). No plumule formation was observed in maize at 150 and 200 mM NaCl.
Table 4
Efficacy of bacterial treatment on plumule length at different NaCl concentrations
Bacterial characterization and identification
Previous studies reported that rhizobacteria phyla, i.e., Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes, are essential for plant growth (Passari et al., 2016; Ashraf et al., 2019). In the present study, BoGl123 was identified and characterized using microscopic, biochemical, and molecular approaches. The results of the microscopic analysis revealed that BoGl123 was rod shaped and gram negative. The BoGl123 16S rRNA gene sequence (1527 nucleotides) showed 97.85% similarity with the Pseudomonas fluorescens strain IAM 12022 and 97.31% similarity with the P. fluorescens strain CCM 2115 (as determined by BLAST search) and was submitted to GenBank under the accession number MT557901. To create a phylogenetic tree for BoGl123, the MEGA X software was used following the Muscle alignment approach (Fig. 3).
Conclusions
Previous studies have demonstrated that PGPRs exert a strong growth-promoting effect on crops, which is likely to evolve during long-term coevolution between the host and microorganisms. The present study showed that B. glabra rhizospheric salinity-tolerant PGPRs improved the growth of wheat and maize under saline conditions. Bacterial inoculation led to promising seed germination and plumule and radicle length results. Such improvements in seed germination parameters enhance the plant hormone production, iron chelator (siderophore) synthesis, solubilization of mineral phosphate, and HCN synthesis, which indicates the beneficial effect of abiotic stress-tolerant bacteria on the growth of wheat and maize seedlings. However, many studies attempted to grow salinity-tolerant crop varieties using transgenic approaches. In transgenic trials, the salt tolerance potential was primarily evaluated in laboratory experiments with a small range of seedlings or mature crops. In most cases, experiments were performed in greenhouse environments where plants were not subjected to conditions prevalent in high-salinity soils (such as pH shift, increased diurnal temperatures, reduced soil moisture, reduced humidity level, and increased prevalence of sodic salts with high levels of metals).
To determine the total productivity of plants in a saline environment, it is necessary to evaluate their salt tolerance potential in the farming field. Measuring productivity in a saline environment is challenging due to the variations in salt concentration in soil and other environmental factors such as soil nutrients, soil temperature, intensity of sunlight during the daytime, and reduced soil water levels. Moreover, the emergence of novel effective bioinoculants has improved crop production and performance in the agricultural sector, which will gradually replace the conventional chemical fertilizer-dependent agriculture techniques. The results of the present study showed that salt-tolerant PGPRs isolated from an abiotic-stress-tolerant plant (B. glabra) rhizosphere enhanced physiological properties (i.e., percent seed germination, root length, shoot length, root weight, shoot weight) of the inoculated seedlings. These PGPRs were involved in the mitigation of the risk pertaining to salinity by enhancing compatible solute deposition and increasing water availabilty. These changes in cellular biochemical reactions resulted in an increased germination rate in susceptible wheat and maize cultivars under saline conditions. Further studies in various field sites with different salt concentrations and extensive examination of molecular processes will eventually contribute to the promising application of halo-tolerant P. fluorescens BoGl123 as biofertilizers to facilitate wheat and maize growth and salt tolerance under saline conditions.