Introduction
Gliomas are prevalent brain tumors, constituting approximately 80% of all malignant tumors in the central nervous system [1]. However, the incidence and mortality of gliomas in East Asia have also been of concern in the last 30 years. Studies have shown that China, the United States, and India are the top three countries with the highest number of cases and deaths from malignant brain tumors of the central nervous system [2, 3]. Glioma patients have a low survival rate, especially in the case of high-grade malignant gliomas [4, 5]. The current strategies for managing glioma commonly involve surgical excision, radiotherapy, and chemotherapy. Nevertheless, the overall rate of successful treatment is still relatively low and patients have a poor prognosis (median survival of less than 2 years) with high chances of recurrence (median progression-free survival of approximately 7 months) [6-8]. Early diagnosis and regular monitoring are crucial for detection of tumor recurrence and implementing effective interventions. Recently, due to the continuous advancement of molecular biology technology and immunotherapy methods, it has become a consensus to elucidate and treat diseases at the molecular level, including in the case of glioma. The screening of novel biomarkers for assisting in the diagnosis and treatment of glioma is of urgent necessity.
Colony-stimulating factors (CSFs) comprise cytokines such as macrophage CSF (M-CSF), granulocyte CSF (G-CSF), granulocyte-macrophage CSF (GM-CSF), and multi-CSF [9, 10]. Colony-stimulating factors stimulated the proliferation, differentiation, and viability of hematopoietic stem cells and specific cell lines in the bone marrow, such as granulocytes, monocytes, and macrophages [11]. However, numerous studies have examined the role of CSFs in tumor initiation, progression, and metastasis [12, 13]. Furthermore, CSFs also regulated tumor progression by affecting the infiltration of immune cells in the tumor microenvironment [14, 15]. CSF3R, also known as G-CSFR, is the gene encoding the receptor for G-CSF. This gene encodes a transmembrane protein called granulocyte colony stimulating factor receptor, which provides proliferative signals for granulocytes and plays a crucial role in their proliferation and differentiation [16, 17]. Initially, research found mutations in CSF3R in patients with severe congenital neutropenia, suggesting that these aberrant mutations are likely early events in leukemia development [18, 19]. Thus, for a long time, most studies primarily focused on the specific mechanisms by which CSF3R regulates leukemia occurrence and development. For example, it was found that 60% of chronic neutrophilic leukemia and atypical (BCR-ABL-negative) chronic myeloid leukemia patients had CSF3R mutations. Transplanting hematopoietic cells with CSF3R mutations into mice led to excessive proliferation of granulocytes in the mice and infiltration of granulocytes into the spleen and liver, resulting in myeloproliferative disorders [20]. Furthermore, research has shown that CSF3R mutations in chronic neutrophilic leukemia patients disrupt a presumed glycosylation site and promote JAK/STAT and MAPK signaling, driving cancer development [21]. These studies indicate the critical role of CSF3R in the progression of hematologic malignancies. However, in recent years, some studies have suggested a close association between CSF3R and the occurrence and progression of various solid tumors. One study found that mice lacking CSF3R showed enhanced macrophage-mediated tumor cell killing in vivo and significantly suppressed tumor growth in mice [22]. Another study suggested that CSF3R deficiency promotes the secretion of interferon γ and interleukin (IL)-17A, reduces IL-10 levels, and inhibits tumor growth [23]. Thus, CSF3R may also play an important regulatory role in solid tumors, although there is currently limited research on its role in solid tumors. In further exploration and study of the relationship between CSF3R and the occurrence, development, and prognosis of solid tumors, this study discovered significant upregulation of CSF3R expression in various solid tumors through pan-cancer analysis. Moreover, abnormal expression of CSF3R was associated with poor prognosis in patients with various solid tumors, especially in gliomas. Therefore, this study focused on analyzing the expression, function, and prognostic relevance of CSF3R in gliomas, aiming to provide valuable insights for their clinical treatment.
Material and methods
Pan cancer analysis of CSF3R
Sangerbox was employed to analyze the expression of CSF3R in diverse tumors and normal tissues, as well as its association with the prognosis of cancer patients.
Data acquisition
This study utilized publicly available databases, including The Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA) datasets, to obtain clinical and transcriptomic data for the samples. Genomic expression data and relevant clinical details were collected from the TCGA dataset through the Genomic Data Sharing Data Portal (GDC; https://portal.gdc.cancer.gov/), and additional clinical data were integrated from the compilation by Michele Ceccarelli [24]. The data from the CGGA datasets were obtained from the CGGA website (http://www.cgga.org.cn/). Patients with available clinical data on overall survival (OS ≥ 30) and World Health Organization (WHO) grading were included. A total of 618 glioma samples were included in the TCGA dataset, while 300 samples were included in the CGGA dataset. The immunohistochemical staining results of CSF3R are derived from the HPA (Human Protein Atlas) database (https://www.proteinatlas.org/).
Survival analysis
This study investigated the potential association between CSF3R expression and OS of glioma patients using patient data extracted from the TCGA and CGGA databases. In addition, univariate and multivariate Cox regression analysis was used to evaluate the clinical and pathologic predictive value of CSF3R. Survival analysis was conducted employing the R package maxstat (version: 0.7-25) and the survfit function from the survival package. The clinical prognosis comparison between the subgroup with low CSF3R expression and the subgroup with high CSF3R expression was conducted using a two-sided log-rank test. Additionally, Kaplan-Meier curve analysis was employed.
Construction and validation of the nomogram model
Utilizing the “rms” R package, a nomogram model was constructed, amalgamating significant prognostic elements (CSF3R expression, WHO grade, age and 1p/19q co-deletion) extracted from both TCGA and CGGA datasets, with the aim of forecasting the OS of glioma patients. To comprehensively evaluate the accuracy and reliability of the model, we further created calibration plots and conducted decision curve analysis (DCA).
Functional enrichment analysis
Based on the median expression level of CSF3R, samples were divided into high-expression and low-expression groups. Gene sets, including Gene Ontology (GO) (c5.go.v2023.2.Hs.symbols) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (c2.cp.kegg_legacy.v2023.2.Hs.symbols), were obtained from the gene set enrichment analysis (GSEA) (https://www.gsea-msigdb.org/gsea/index.jsp). Enrichment pathways were filtered using GSVA with the criteria of log fold change (logfc) ≥ 0.3 and p < 0.05.
Immunological features related analysis
The enrichment score for each sample was calculated using single-sample gene set enrichment analysis (ssGSEA) across 29 distinct “immune profiles” encompassing a variety of immune cell types, functions, and immune checkpoint genes [25]. Subsequently, samples were sorted according to CSF3R expression, and heat maps were generated using the corresponding ssGSEA scores. For a comprehensive assessment of the glioma tumor microenvironment (TME), the ESTIMATE algorithm was utilized to assess immune score, stromal score, tumor purity, and the presence of an estimate score [26]. The infiltration levels of various tumor-infiltrating immune cells (TIICs) in the tumor microenvironment of each sample were assessed using the deconvolution method based on linear support vector regression (CIBERSORT) [27]. Subsequently, we compared the infiltration levels of TIICs among different subgroups and assessed for potential differences.
Immunotherapy response analysis
Based on RNA expression levels, we assessed each patient’s clinical response to immunotherapy using the Tumor Immune Dysfunction and Exclusion (TIDE) algorithm (http://tide.dfci.harvard.edu/) and obtained the TIDE scores for each patient. Additionally, we compared the differences in expression of 27 immune checkpoints between the CSF3R low-expression group and the high-expression group [28, 29].
Single-cell analysis
The single-cell analysis for CSF3R was conducted using Tumor Immune Single-cell Hub 2 (TISCH2) (http://tisch.comp-genomics.org/home/) [30]. Quantitative and visual analysis of CSF3R expression levels in 17 cell types was performed through heat maps, scatter plots, and violin plots. CSF3R, major lineage cell type annotation, and glioma cancer type were used as analysis parameters.
DepMap
The DepMap database (https://depmap.org/portal/) provides a comprehensive resource for genome-wide CRISPR-Cas9 gene knockout screens and gene expression data across multiple cell lines. To analyze gene expression, the DepMap database was utilized to obtain target gene expression levels across multiple cell lines. To assess the impact of gene knockout on cell survival, we performed gene-specific analysis using the CRISPR-Cas9 knockout data from the DepMap database and obtained information on cell line viability following knockout of the target gene.
Statistical analysis
All statistical analyses in this study were conducted using the R programming language (version 4.3.0, https://www.r-project.org/) and SPSS 29.0 (IBM Corp., Armonk, NY, USA, Version 29.0). Various statistical methods were applied to analyze the data, including Student’s t-test, Pearson correlation analysis, and the log-rank test. A significance level of p < 0.05 was considered to indicate statistical significance.
Results
CSF3R is highly expressed in gliomas
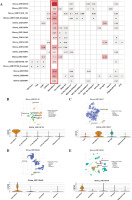
By extracting the expression of CSF3R from the TCGA TARGET GTEx dataset in the UCSC database, we observed distinct expression patterns of CSF3R across multiple tumors (Fig. 1A). Elevated expression of CSF3R was associated with poor OS and disease-specific survival (DSS) in various cancer patients, particularly in glioma patients (Fig. 1B and Supplementary Fig. S1). Subsequently, a comparison of CSF3R expression was performed between 163 glioblastoma (GBM) samples and 207 normal samples, along with 518 low-grade glioma (LGG) samples and 207 normal samples. These results indicated notable upregulation of CSF3R in GBM and LGG samples when compared to normal samples (Fig. 1C, D). The immunohistochemical analysis of data from the HPA revealed a significant increase in CSF3R expression within tumor tissues (Fig. 1E-G). These results indicated high expression of CSF3R in gliomas, suggesting its potential role as an oncogene.
Fig. 1
Expression of CSF3R in pan-cancer and in glioma. A) Expression of CSF3R between the 34 cancers and normal tissues in unpaired sample analysis. B) Expression of CSF3R is associated with poorer overall survival (OS) in tumor patients. C, D) Expression of CSF3R in low-grade glioma (LGG) and GBM compared with normal tissues. E-G) Expression of CSF3R in LGG and glioblastoma (GBM) from HPA
Associations of CSF3R expression with molecular and clinical characteristics in gliomas
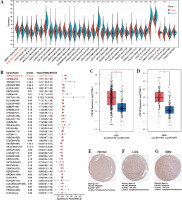
Extensive studies have confirmed the associations of malignant gliomas with isocitrate dehydrogenase (IDH) mutation and 1p/19q co-deletion status [31]. Patients with different levels of CSF3R expression exhibited distinct clinical and pathological characteristics. The expression of CSF3R was elevated in patients with IDH wildtype status and 1p/19q non-codeletion status in the TCGA dataset. Moreover, CSF3R expression increased significantly with the elevation of WHO grade levels (Fig. 2A). These results further validated the CGGA database (Fig. 2B) and suggested that the elevated expression levels of CSF3R are positively correlated with the increased malignancy of gliomas. Subsequently, we analyzed the expression of CSF3R in various glioma cell lines using the DepMap database. As illustrated in Figure 2C, the results revealed a prominent upregulation of CSF3R in various glioma cell lines, including U87MG, 8MGBA, YH13, LN229, and KALS1. Additionally, DepMap was utilized to predict the regulatory impact of CSF3R on glioma cell survival, elucidating whether it promotes or inhibits this process. The results showed a significant reduction in the viability of the majority of cells upon CRISPR-mediated knockout of CSF3R in glioma cell lines (Fig. 2D).
Fig. 2
Expression of CSF3R is associated with the clinical characteristics of gliomas as well as the survival rate of glioma cells. A, B) Expression of CSF3R is significantly correlated with IDH mutation, 1p/19q codeletion status, and WHO grading, with no apparent correlation with age. C) Expression profile of CSF3R across different glioma cell lines, with log2 (TPM+1) values above 0 indicating high expression. D) Representation of CRISPR-mediated knockout of CSF3R in glioma cell lines, depicting cell viability. A score below 0 signifies CSF3R gene knockout and reduced cell survival rate
CSF3R as an independent prognostic factor and in association with glioma patient survival
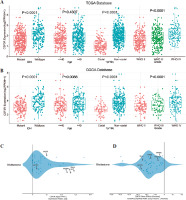
The above results suggested that CSF3R was highly expressed in gliomas and may be associated with the malignant progression of gliomas. Therefore, Kaplan-Meier survival analysis was used to further assess the relationship between the expression of CSF3R and the prognosis of glioma patients. The results indicated that in the TCGA database, patients with elevated CSF3R expression had a lower OS (median survival time: 1012 days) compared to patients with lower CSF3R expression (median survival time: 2869 days) (Fig. 3A). Consistent results were observed in the CGGA database (Fig. 3B). ROC analysis demonstrated that using CSF3R to predict the 1-year, 3-year, and 5-year survival rates of patients has great utility (Fig. 3C, D). Subse- quently, Cox regression analysis revealed that CSF3R expression serves as an independent prognostic factor, independent from known prognostic factors including WHO grade, age, IDH mutation and 1p/19q co-deletion (Fig. 3E, F). The above results indicated CSF3R as an independent prognostic factor in gliomas.
Fig. 3
Potential prediction of CSF3R as a prognostic factor for glioma. A, B) Kaplan-Meier survival analysis showed that in the TCGA and CGGA databases, patients with high CSF3R expression had lower overall survival (OS). C, D) ROC curve for the predictive efficiency of Kaplan-Meier survival curves. E, F) Univariate and multivariate analysis indicated that CSF3R could be used as an independent prognostic factor in glioma

Construction and validation of a nomogram-based model of CSF3R expression
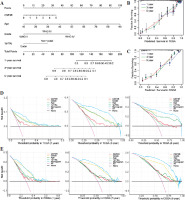
In order to more comprehensively assess the value of CSF3R in predicting patient prognosis, this study constructed a nomogram model to assess the prognostic ability of CSF3R. Based on the nomogram model, it can be inferred that patients with high expression of CSF3R have a poorer prognosis compared to those with low expression of CSF3R (Fig. 4A). The calibration curves for both the TCGA and CGGA cohorts demonstrated a high level of accuracy, with a C-index of 0.857 for TCGA and 0.760 for CGGA (Fig. 4B, C). These results suggested the reliability of using CSF3R as a predictor for 1-year, 3-year, and 5-year overall survival in glioma patients. According to the DCA curves constructed in this study, we observed that the expression of CSF3R demonstrates favorable practicality in both the TCGA and CGGA cohorts. Additionally, compared to the individual use of CSF3R expression, patient age, WHO grade, and chromosomal 1p/19q status for predicting patient OS, the nomogram demonstrated greater advantages (Fig. 4D, E).
Fig. 4
Nomogram and decision curve analysis (DCA) curves. A) The nomogram was constructed with WHO grade, age, and CSF3R expression in the TCGA cohort. B, C) The calibration plots: verify the accuracy of the predicted 1-, 3- and 5-year overall survival (OS) in the nomogram. D, E) DCA curves: CSF3R, grade, age and 1p19q curves represent their own prognostic value. Nomogram curve represents the synthetical prognostic value of CSF3R, grade, age and 1p19q. No curve represents no prognostic value. All curve represents the theoretical best prognostic value
CSF3R involvement in immune responses and signaling pathways in gliomas
To explore the biological roles of CSF3R, the samples from TCGA and CGGA datasets were categorized into high and low expression clusters according to the median CSF3R expression level. Subsequently, we observed a significant correlation between elevated CSF3R expression and immune response and inflammatory reaction, including primary immunodeficiency, intestinal immune network for immunoglobulin A (IgA) production, cytokine receptor interaction, mast cell mediated immunity, pattern recognition receptor activity, interleukin 3 mediated signaling pathway, canonical inflammasome complex, leukocyte degranulation, and neutrophil activation involved in the immune response (Fig. 5). These results suggested that CSF3R in gliomas may play a crucial role in regulating immune responses.
The relationship between CSF3R and immune characteristics in gliomas
The previous results have suggested that CSF3R may be associated with various immune-related pathways. In the subsequent analyses, similar results were obtained. The expression of CSF3R exhibited robust associations with several immune signatures, such as macrophages, mast cells, MHC class I, checkpoints, T cell co-stimulation/co-inhibition, Th1/Th2 cells and B cells, and others (Fig. 6A, B and Supplementary Fig. 2A, B). Additionally, the high CSF3R expression subgroup exhibited notably increased ImmuneScore, StromalScore, and Estimatescore, while showing reduced TumorPurity (Fig. 6C, D and Supplementary Fig. 2C, D). We further investigated the correlation between CSF3R and tumor-infiltrating immune cells; however the results indicated in TCGA and CGGA databases a positive correlation between the expression level of CSF3R and the infiltration levels of monocytes and M2 macrophages (Fig. 6E-G and Supplementary Fig. 2E-G). Subsequently, we gathered 27 established immune checkpoints from published research and observed significant differences in the expression of multiple immune checkpoints across samples with different levels of CSF3R (Fig. 6H and Supplementary Fig. 2H). These results further underscored the crucial role of CSF3R in modulating the immune response within the glioma microenvironment.
Fig. 6
Association of CSF3R with immune regulation and immune treatment in glioma. A, B) Heatmaps of enrichment scores based on “immune signatures” in The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) datasets. C, D) Correlation of CSF3R with ImmuneScore in TCGA and CGGA databases. E) Comparison of TIICs between the low and high CSF3R expression groups in TCGA and CGGA databases. F, G) Correlation of CSF3R with monocytes and M2 macrophages in TCGA and CGGA databases. H) Difference of immune checkpoint expression in low and high CSF3R subgroups in TCGA. I-L) TIDE, T cell dysfunction, MSI and T cell exclusion score in low and high CSF3R subgroups in TCGA
The TIDE score was used to evaluate the efficacy of immunotherapy and the potential for tumor immune escape based on the gene expression profile. A higher TIDE score indicates a stronger ability of the tumor to evade the immune system, leading to a less favorable response to immune checkpoint inhibition [32]. In this study, patients with high expression of CSF3R exhibited significantly higher TIDE scores compared to the low-expression group, indicating a stronger capability of tumor immune escape in the CSF3R high-expression group (Fig. 6I). Moreover, patients in the high-expression group of CSF3R exhibited increased microsatellite instability (MSI), elevated T cell dysfunction scores, and reduced exclusion scores (Fig. 6J-L). These findings indicated that patients with high CSF3R expression are more prone to developing resistance during immune checkpoint inhibition therapy, thereby promoting tumor escape and malignant progression.
Expression of CSF3R on different cells in the tumor microenvironment
The aforementioned analysis outcomes implied a potential pivotal role of CSF3R in the tumor immune response. To ascertain the expression profiles of CSF3R across distinct cellular components within the tumor microenvironment, we proceeded to perform single-cell analysis of CSF3R utilizing the TISCH network tool. The range of CSF3R expression in a variety of cell types, including immune cells, stromal cells, malignant cells, and functional cells, is shown visually in Figure 7A. Notably, CSF3R exhibited predominant expression in monocytes/macrophages. In the GSE102130 dataset, which utilized single-cell RNA sequencing to analyze 4058 K27M-mutant glioma cells, CSF3R expression was primarily observed in monocytes and macrophages within the GBM microenvironment (Fig. 7B). Similarly, CSF3R was predominantly expressed in monocytes and macrophages in the GSE135437, GSE139448, and GSE138794 datasets (Fig. 7C-E). These results further emphasized the association between CSF3R and monocytes/macrophages in the tumor microenvironment. Therefore, the above information suggested that CSF3R may impact the immune processes of gliomas by regulating monocytes/macrophages.
Discussion
Glioma is a common type of brain tumor, and its etiology is elusive. To a large extent, gliomas are still incurable at present [33]. Research suggests that factors such as radiation exposure, viral infections, and genetic predisposition may contribute to glioma development [34, 35]. In addition to these factors, an increasing body of evidence indicates that glioma pathogenesis is closely associated with dysregulated gene expression, genetic mutations, and aberrant signaling pathway activation [36-38]. For instance, one study found that the expression of protein arginine methyltransferases is upregulated in gliomas, which promotes the transcription of the CDC20 factor by activating histone methylation marks. CDC20, upon binding to CDKN1B, promotes the ubiquitination of CDKN1B, leading to its decreased stability and thereby accelerating glioma cell proliferation [39]. Moreover, research has shown that the EGFR/SRC/ERK pathway promotes glioma cell proliferation and migration by upregulating the expression of YTHDF2 [40]. Thus, investigating the molecular biology of glioma pathogenesis may help elucidate the underlying mechanisms and facilitate clinical diagnosis and treatment of gliomas.
Here, we detected high expression of CSF3R in gliomas, which is associated with the malignant progression of gliomas and poor prognosis in patients. Categorizing patients into high and low expression groups based on CSF3R levels, survival analysis indicated that individuals with elevated CSF3R expression experienced poorer OS. Furthermore, both univariate and multivariate Cox analyses suggested that CSF3R exhibited unique advantages in predicting 1-, 3-, and 5-year OS in glioma patients. The nomogram constructed in this study, which incorporated factors such as CSF3R expression, demonstrated significant clinical value in predicting the OS of glioma patients. These results underscored the potential of CSF3R as a prognostic marker in gliomas.
Subsequently, to further investigate the mechanisms by which CSF3R controls the malignant progression of gliomas and patient prognosis, this study employed the GSVA algorithm to explore the potential biological functions of CSF3R. The results indicated that CSF3R is involved in the regulation of various immune-related functions. The status and proportion of immune cells in the tumor microenvironment are closely associated with the malignant progression of the tumor. The expression of CSF3R is associated with the abundance of various immune cells, and notably, we observed a significant correlation between CSF3R expression and the infiltration level of M2-type macrophages. The single-cell analysis also revealed that within the tumor microenvironment of gliomas, CSF3R was predominantly expressed in monocytes/macrophages. Monocytes/macrophages derived from bone marrow and circulated in the bloodstream play a crucial role in immune response reactions [41, 42]. Monocytes have been found to migrate towards inflammatory or injured sites and differentiate into macrophages [43]. As the primary responders of the immune system, macrophages engaged in phagocytosis and digestion of dead cells, cellular debris, and pathogens, contributing to the clearance of infections and promoting tissue healing [44, 45]. Additionally, macrophages produced and released various cytokines to activate lymphocytes or other immune cells, which play a pivotal role in coordinating immune responses [46, 47]. Recent studies have shown that brain-resident macrophages are the primary immune cell populations involved in the central nervous system immune defense of glioma patients [48-50]. Meanwhile, macrophages are closely associated with the progression of gliomas. A recent study revealed that lactate dehydrogenase A (LDHA)-directed extracellular signal-regulated kinase (ERK) signaling serves as a transcriptional coactivator for YAP/STAT3 in glioblastoma cells, upregulating CCL2 and CCL7, which recruit macrophages into the tumor microenvironment. Subsequently, infiltrating macrophages produce extracellular vesicles containing LDHA, promoting glycolysis, proliferation, and survival of glioma cells. Furthermore, it has been demonstrated that PTEN-deficient glioma cells can increase the secretion of galectin-9 through the AKT-GSK3β-IRF1 pathway. Elevated galectin-9 activates Tim-3 in macrophages and downstream pathways to promote M2 polarization, leading to increased secretion of VEGFA, thereby stimulating angiogenesis and facilitating glioma growth [51]. As described above, in this study, we found a positive correlation between the expression level of CSF3R and the infiltration level of M2 macrophages [52]. Therefore, we speculate that CSF3R may influence the progression of gliomas by modulating the polarization of macrophages towards the M2 phenotype.
During the growth of tumors, inhibitory signals associated with the immune response, also known as checkpoints (including PD-1/PD-L1, CTLA4, and TIGIT, among others), are activated. These signals suppressed the ability of the immune system to clear tumor cells and promoted the malignant progression of tumors [53-56]. In recent years, immune checkpoint blockade therapy, aimed at activating the immune response, has demonstrated significant effectiveness in treating various tumors, including gliomas [57, 58]. A representative study revealed that the combination of anti-PD-1 therapy with anti-TIGIT therapy results in an increased population of effector T cells in mice with gliomas. This combination leads to a reduction in suppressive Tregs and tumor-infiltrating dendritic cells, thereby inhibiting the malignant progression of tumors in mice and extending their survival [59]. Additionally, glioma patients with high PD-L1 expression had a higher proportion of M2-type macrophages [60]. However, anti-PD-1 therapy promoted the polarization of M1-type macrophages in the glioma microenvironment, thereby inhibiting glioma progression. This effect was observed in mice lacking CD8+ T cells in vivo [61]. Thus, it is evident that the strategy of immune checkpoint blockade holds significant value in the treatment of gliomas. Interestingly, this study revealed a correlation between CSF3R levels and the expression of various immune checkpoint markers. Patients with elevated CSF3R expression demonstrated a diminished response to immune checkpoint blockade therapy, indicating an increased probability of tumor immune evasion (lower TIDE scores in patients with high CSF3R expression). The aforementioned results suggested that CSF3R may serve as a potential therapeutic target, especially in combination with immune checkpoint blockade therapy. As mentioned previously, our study has identified CSF3R as a potential target for monocytes/macrophages. Consequently, this research postulates that CSF3R may influence the effectiveness of immunotherapy mediated by monocytes/macrophages, thereby impeding the progression of gliomas.
In our study, there are also several limitations that should be acknowledged. The analysis of CSF3R in glioma was based on public databases. Although an adequate sample size and good homogeneity were ensured, inherent biases in retrospective studies cannot be entirely avoided. Furthermore, demonstrating the value of CSF3R in glioma treatment based solely on bioinformatic analyses is inadequate. It is crucial to conduct experimental validations to further explore the mechanisms and clinical significance of CSF3R in glioma treatment, facilitating a more in-depth and specific assessment.
Conclusions
In summary, this study reveals a significant association between CSF3R and the clinical pathological features, adverse prognosis, and polarization of macrophages in the tumor microenvironment of gliomas. Therefore, CSF3R has the potential to serve as a prognostic marker for gliomas and as a specific therapeutic target, which may facilitate the development of targeted therapeutic drugs for gliomas and enhance clinical treatment efficacy.