Introduction
Psoriasis is a serious inflammatory skin disorder with a worldwide incidence of 1–3% [1, 2]. It is prone to recurrent attacks and brings great distress to patients. Psoriasis is an immune-mediated chronic inflammatory skin disease. The pathology is distinguished by increased growth of keratinocytes and parakeratosis. Systemic and environmental exposures, namely interleukin 17 (IL-17) and interleukin 23 (IL-23), are considered key drivers of disease [3], cytokines namely interleukin 16 (IL-6) and tumor necrosis factor-α (TNF-α) are functionally engaged in the pathogenesis [4].
Autophagy is a lysosome-mediated catabolic pathway widely present in eukaryotic cells. Under conditions such as starvation, stress, or growth factor depletion, autophagy will be rapidly induced and started to degrade proteins, and damaged organelles (such as mitochondria, etc.) and remove intracellular pathogens, etc., to generate new substances and energy to maintain cells’ survival. Different from pyroptosis [5], autophagy does not directly cause an inflammatory response but has a significant impact on differentiation and proliferation, metabolism and cell growth, and inflammation regulation [6]. Autophagy is an evolutionarily conserved process for preserving cellular homeostasis and is the self-degradation of defective organelles, misfolded or aggregated proteins, and certain aging proteins [7]. Autophagy is identical to autoimmune disorders, namely systemic lupus erythematosus, psoriasis, rheumatoid arthritis, etc. Significantly enhanced expression, activation of phosphoinositide-3 kinase/protein kinase B/mammalian rapamycin protein inhibition of autophagy has a positive therapeutic effect on IL-17a-mediated psoriasis. The expression of the autophagy-related gene (ARG) Sequestrum 1 was significantly enhanced in psoriatic lesions, and the activation of phosphoinositide-3 kinase/protein kinase B/mammalian rapamycin inhibited the effect of autophagy on IL-17a-mediated Leading psoriasis has a positive therapeutic effect.
Recently, with the progress and wide application of gene sequencing technology, bioinformatics analysis based on sequencing data can efficiently process a large amount of sample data and provide valuable information about diseases. Wang et al. [8] obtained chemokine (C-X-C type) ligand 9 (CXCL9), small proline-rich protein 1B (SPRR1B), transglutaminase 6 (TGM6), and S100 calcium-binding protein based on bioinformatics methods. A9 (S100A9) 4 key genes in the progression of psoriasis, among which CXCL9 has a strong chemotactic effect on monocytes, neutrophils, and T lymphocytes that are important in the inflammatory process. SPRR1B is identified as a potential biomarker for psoriasis [9]; TGM6 affects the differentiation of keratinocytes, and S100A9 is important in the amplification of psoriasis inflammatory process due to chemotaxis, adhesion, and degranulation of neutrophils play a role. However, the use of bioinformatics to analyse psoriasis autophagy-related key genes (Hub) has rarely been reported in the literature. In this study, the sequencing data of psoriasis in the GEO database were selected for relevant bioinformatics analysis, and the possible related Hub genes and possible mechanism pathways were explored.
Material and methods
Data source
The Human Autophagy Dataset (http://www.autophagy.lu/index.html) provided 222 genes in total. The GEO dataset (www.ncbi.nlm.nih.gov/geo/) is a gene database generated by the National Center for Biotechnology Information (NCBI), which collects gene expression data submitted by research institutions around the world, and is currently the most widely used gene database [10]. The expression profile of the GSE78097 dataset was downloaded from this database through the GEOquery package. GSE78097 is located in GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, which comprises 27 psoriasis and 6 normal skin tissue samples.
Differential expression analysis of autophagy-associated genes
From the GSE78097 dataset, a normalized expression matrix for microarray data was obtained. The probes are then annotated using the dataset’s annotation files. Principal component analysis (PCA) was utilized to confirm the reproducibility of the information in GSE78097. The R software tool was utilized to recognize differentially expressed autophagy-related genes. Absolute fold-change value > 1.5 and p-value < 0.05 were used to determine which genes were differentially expressed. Construction of heatmaps, boxplots, and volcano plots was performed using the “ggplot2” and “heatmap” R software’ packages.
Correlation and PPI analyses of differentially expressed autophagy-associated genes
The Cytoscape software (version 3.8.1) and STRING dataset (https://string-db.org/) were utilized to examine PPI analyses of differentially expressed autophagy-associated genes [11]. Utilizing Spearman correlation, the “corrplot” package of the R software was employed to recognize the correlation analysis of differentially expressed autophagy-associated genes.
KEGG and GO enrichment analysis
GO and KEGG pathway enrichment analyses were conducted utilizing the “GO plot” package in the R software [12]. Biological processes (BP), molecular functions (MF), and cellular elements (CC) were all included in the GO study.
Results
Screening of differentially expressed autophagy-associated genes
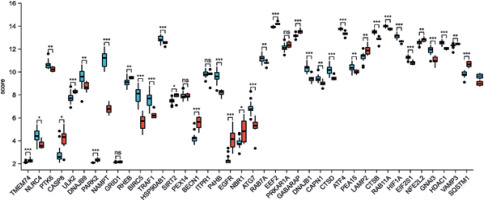
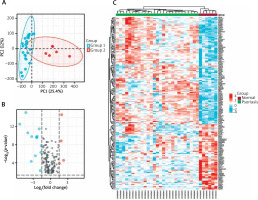
We used principal component analysis (PCA) to evaluate the group’s data reproducibility, and the results revealed that the GSE78097 data had good reproducibility (Figure 1 A). Next, the expression of 222 kinds of autophagy was examined. Absolute fold-change value > 1.5 and p-value < 0.05 allowed us to identify 156 genes associated with autophagy. The volcano plot shows that 156 autophagy-related genes were differentially expressed among psoriasis and healthy groups (Figures 1 B, C). Additionally, the box plot also revealed the expression patterns of the top 40 differential genes among these 156 differentially expressed autophagy-related genes among healthy and psoriasis specimens (Figure 2).
Figure 1
Autophagy-associated genes differentially expressed in healthy and psoriasis specimens. A – PCA of GSE78097; B – Volcano plot of 222 differentially expressed autophagy-associated genes. Blue spots display remarkably down-regulated genes, and red spots display remarkably up-regulated genes; C – Heatmap of 156 differentially expressed autophagyassociated genes in psoriasis and normal specimens
Correlation and PPI analyses of differential expression of autophagy-associated genes
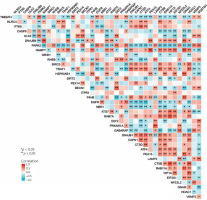
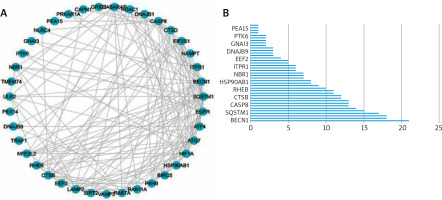
We utilized a PPI study to recognize the relationships between differentially expressed autophagy-associated genes. The outcomes demonstrated associations between these autophagy-related genes (Figure 3 A), and the interaction numbers of 13 genes that were meaningful among them were shown (Figure 3 B).
Figure 3
Protein-protein interactions (PPIs) of 40 differentially expressed autophagy-associated genes. A – PPI network diagram between 40 differentially expressed autophagy-associated genes; B – interaction numbers for all differentially expressed autophagy-associated genes
We also carried out a correlation analysis to investigate the expression connection of these autophagy-related genes, and Figure 4 depicts the relationships between the 40 differentially expressed autophagy-associated genes in the GSE78097 database.
KEGG and GO enrichment analysis of differentially expressed autophagy-associated genes
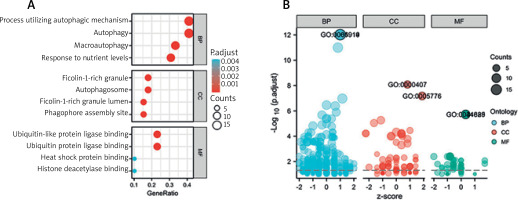
We utilized R software to conduct KEGG and GO enrichment analysis to investigate the possible biological roles of these differentially expressed autophagy-associated genes. The outcomes of the Gene ontology study showed that the potential biological functions mainly include macroautophagy, phagocyte assembly site, autophagosome, ubiquitin-like protein ligase binding, lumen rich in ficolin-1 particles, ubiquitin protein ligase binding, group Protein deacetylase binding and heat shock protein binding (Figures 5 A, B, Table 1). The outcomes of KEGG analysis revealed that the potential biological functions mainly include apoptosis, protein processing in mitochondria, endoplasmic reticulum, NOD-like receptor signalling pathway, oestrogen signalling pathway, Shigellosis, pathways of neurodegeneration and apoptosis, etc. (Figure 6, Table 2).
Table 1
Details of GO enrichment analysis
Table 2
Detailed results of KEGG analysis
Discussion
Psoriasis is a serious relapsing inflammatory skin disorder with a relatively significant clinical rate. Its pathological features are excessive cell proliferation, abnormal differentiation, and parakeratosis leading to epidermal keratinization, inflammatory cell infiltration between the epidermis and dermis, and expansion, hyperplasia, and tortuosity of microvessels in the papillary dermis [13]. Currently, only a few investigations have explored the association between psoriasis and autophagy-related genes. For example, the loss of autophagy-related 16-like protein 1 can induce cell death, tissue damage, and inflammation, and its polymorphism is associated with psoriasis; inhibition of autophagy can stimulate keratinocytes to produce autonomous cell proliferation inhibition [14]. Therefore, we need further research and analysis to provide better references for the clinical treatment and management of psoriasis.
Through the method of bioinformatics analysis, this study finally obtained the top 40 differential genes. Among these genes, we can find that some related studies have been carried out, such as the heat shock protein gene is related to the generation of psoriasis fatigue, supporting the hypothesis that downregulation of the innate immune response affects fatigue [15]; keratinocytes are important cellular targets for IL-17A-mediated effects in psoriasis, and HSP90 is crucial for IL-17A-mediated signalling, HSP90 inhibitors RGRN-305 significantly reduced the expression of many pro-inflammatory genes among those induced by IL-17A and TNF-α, namely the psoriasis-related genes CCL20, IL23A, NFKBIZ, IL36G [16]; the genes of CASP7 and CASP8 in a Han population in Northeast China Polymorphisms were significantly associated with psoriasis vulgaris, suggesting a significant connection among psoriasis vulgaris and caspase genes. CASP7 and CASP8 SNPs may be new biomarkers for risk prevention and assessment of psoriasis vulgaris [17, 18]. A clinical study included 1825 patients receiving ixekizumab treatment. Research has shown that consistent with the clinical results of the overall population, all self-identified racial groups showed rapid improvement in psoriasis area and severity index at week 12, although the response of Native American/Alaska Indigenous patients was slightly slower. There are differences in reporting results and quality of life assessments among different racial groups, especially among black/African American and Native American/Alaska Indigenous patients [19]. Keratinocytes negatively regulate p62 expression through autophagy, which is important for preventing excessive inflammation and inducing human inflammation in psoriasis. Catheterin in keratinocytes is critical. However, there is still a lack of research on other autophagy-related genes in psoriasis, and more exploration and analysis are needed.
KEGG and GO enrichment study was utilized to examine the possible biological roles of these differentially expressed autophagy-associated genes. The outcomes showed that autophagy, apoptosis, mitochondria, protein synthesis, etc., may significantly contribute to the progression and occurrence of psoriasis. This also reflects the significant role of autophagy in the pathogenesis of psoriasis. There are also related studies in the past, such as PSORI-CM02 induces autophagy by inhibiting the PI3K/Akt/mTOR pathway, and improves psoriasis in vitro and in vivo [20]. Anthraquinone, an anti-psoriasis drug, gathers in keratinocyte mitochondria, vanish mitochondrial membrane ability, and induces apoptosis through a route dependent on respiratory competent mitochondria. In HaCaT cells, nitidine chloride promotes mitochondria-dependent apoptosis and S-phase cell cycle arrest and improves psoriasis-like mouse model skin damage [21].
This study achieved the expected results by using bioinformatics methods, but there are still some limitations: 1) The difference in age distribution between patients and controls in the sample makes it impossible to distinguish the differences in autophagy levels between patients of different ages; 2) The research and analysis data are all taken from the GEO database. The accuracy of the data and the limited sample size may cause deviations in the research results, which need to be verified in subsequent basic experiments and clinical practice.