Introduction
Ovarian cancer (OC) is one of the most common cancers in women worldwide, and a leading cause of death among gynecological malignancies [1]. The incidence of OC among Tunisian women was approximately 4/100,000, with an estimated 93 new cases diagnosed annually [2]. The pathogenesis of OC is not well characterized. In fact, most patients at diagnosis present with advanced stage of disease, and its treatment is refractory to chemotherapy and surgery [3]. The lack of available sensitive and specific screening tests, along with the identification of predictive and prognostic factors, contributes to the aggravation of OC [4, 5].
It has been established that an inflammatory reaction is the underlying cause in the development of many types of cancers [6]. Molecular mechanisms causing inflammation-associated cancer include DNA damage disruption of immune response and alternation of the tumor microenvironment, which are all closely related to the disequilibrium of inflammatory cytokines [7, 8].
In this context, few studies have focused on identifying novel biomarkers associated with OC susceptibility, which in turn may facilitate diagnosis, and possibly clinical management [9-11]. Moreover, previous studies have shown that the growth and progression of OC result from a state of immunosuppression, which facilitates tumor outgrowth protecting tumor from immune system controlled by cytokines [12, 13]. Given their role in initiating and sustaining immunologic and inflammatory processes, a strong correlation between chronic inflammation, cytokine expression, and OC development have been reported [14, 15].
Transforming growth factor β (TGF-β1) is a multi-functional anti-inflammatory cytokine involved in the control of cell cycle and plays a role in carcinogenesis through regulating cellular growth, differentiation, adhesion, migration, and apoptosis [16, 17].
TGF-β1 reportedly affects early stage of cancer and promotes growth of cancerous cells in later stage [18]. The immunosuppressive role of TGF-β1 correlates with its overexpression in later stages of cancer and is dependent on its level of expression by DNA variation in its promoter or regulatory sequences [19, 20]. The TGF-β1 gene is located on chromosome 19, and several polymorphic variants in the promoter region of TGF-β1 gene have been shown to cooperate in modulating transcription factor binding and with the development of OC [21, 22]. Despite the importance of TGF-β1 in carcinogenesis, few studies evaluated the association of TGF-β1 gene polymorphisms and the risk of OC [22-25].
Interleukin 6 (IL-6), in contrast, is a pro-inflammatory cytokine, with a critical role in inhibition of apoptosis, stimulation of cellular proliferation and angiogenesis, enhancing the invasive and metastatic potential of several cancers [26]. The human IL-6 gene is mapped to chromosome 7p21-24, composed of four introns and five exons, with an upstream promoter containing 303bp [27]. Previous studies demonstrated the involvement of this cytokine in different physiological and pathophysiological processes, such as bone metabolism, synthesis of CRP, and carcinogenesis progression [28, 29].
The IL-6 polymorphism has been regarded as a crucial modulator in pathogenesis of various types of cancer [30-34]. However, allelic distributions of various polymorphisms in IL-6 gene could vary geographically and ethnically, thus leading to discordant findings between these polymorphisms and cancer risk. With respect to the gynecological cancers, it has been reported that increased IL-6 protein levels are associated with malignant ovarian tumors, endometriosis, and cervical cancer [35, 36]. Therefore, in vitro studies have demonstrated that various ovarian carcinoma cell lines produce IL-6, which is thought to be involved in ovarian carcinogenesis by mediating host immune responses to the disease [29, 37, 38].
The purpose of this pilot study was to investigate the contribution of TGF-β1 common variants (rs1800470, rs1800472, and rs1800469) and IL-6 polymorphisms (rs1880242, rs2069827, rs1800797 rs1800796, rs1800795, rs1474348, rs1474347, and rs2069845) to OC susceptibility among Tunisian population.
Material and methods
Study subjects
This case-control study was performed between 2012 and 2013 at Salah Azeiz Oncology Institute (SAI), Tunis, Tunisia. Study subjects consisted of 71 patients with histologically confirmed OC and 74 age-matched healthy controls, with no malignancy, drug allergy, hypertension, diabetes, or cardiovascular disease. Clinical data was collected through self-reported questionnaires, and tumor staging was evaluated according to the International Federation of Gynecology and Obstetrics (FIGO) classification (www.figo.org). Study subjects were from North Tunisia, and all women signed an informed consent to participate in the study. The study was approved by the local ethic committee of Salah Azeiz Oncology Institute.
DNA extraction
Blood samples were taken from all participants using EDTA-containing tube for total genomic DNA extraction shortly before radiation therapy or chemotherapy. Genomic DNA was extracted using QIAamp DNA blood mini kit, according to manufacturer’s instruction (Qiagen GmbH, Hilden, Germany).
Genotyping of TGF-β1 and IL-6
SNPs in TGF-β1 and IL-6 genes were selected, based on minor allele frequency (MAF) of > 5% in Caucasians. TGF-β1 (rs1800470, rs1800472, and rs1800469) and IL-6 (rs1880242, rs2069827, rs1800797 rs1800796, rs1800795, rs1474348, rs1474347, and rs2069845) genotyping was performed by allelic (VIC- and FAM-labeled) discrimination method. TaqMan assays, as assay-on-demand, were ordered from Applied Biosystems (Foster City, CA, USA). The reaction was performed in a 6 µl volume on StepOne/StepOne Plus real-time PCR systems, according to manufacturer’s instructions (Applied Biosystems, Foster City, CA, USA). Replicate blinded quality control samples were included to assess the reproducibility of genotyping reaction, with concordance of > 99%. Additional quality control measures with direct DNA re-sequencing of patients (n = 40) and controls (n = 40) specimens (ABI 3130_l Genetic Analyzer; Applied Biosystems) were applied. A genotyping call rate exceeded 99%, with no significant differences between cases and control samples.
Statistical analysis
Statistical analysis was performed on SPSS v. 21.0 (SPSS Inc., Chicago, IL, USA). Data was expressed as percentages of total (categorical variables) or as mean ±SD (continuous variables). Student’s t-test was used to determine differences in means, and Pearson-V2 or Fisher’s exact tests were used to assess inter-group significance. Allele frequencies were calculated by the gene-counting method, and each polymorphism was tested for Hardy-Weinberg equilibrium with c2 goodness-of-fit test using SNPStats software (http://bioinfo.iconcologia.net/snpstats). LD analysis and haplotypes reconstruction was performed with Haploview 4.1 (http://www.broad.mit.edu/mpg/haploview). Bonferroni multiple comparison correction method was employed in calculating the corrected P (Pc) value: Pc = 1 – (1 – P)n, where n = number of comparisons. Logistic regression analysis was performed to determine odds ratios (OR) and 95% confidence intervals (95% CI) associated with OC risk, taking the control women as the reference group. Statistical significance was set at p < 0.05.
Results
Study subjects
Demographic and clinical characteristics of cases and controls are described in Table 1. Median age was 51 years. Among OC patients, 36 (50.70%) were older than 50 years and 43 (60.60%) were post-menopausal. Diagnoses of OC confirmed by histology according to FIGO revealed 10 patients (14.10%) with stage I, 25 (35.20%) with stage II, 30 (42.30%) with stage III, and 6 cases (8.50%) with stage IV. The majority of histological types found included serious papillary cell carcinoma (49 patients, 69.00%), followed by endometrioid (14 cases, 19.70%), mixed/others (7, 9.90%), and mucinous (1 patient, 1.40%).
Table 1
Characteristics of the study participants
TGF-β1 and IL-6 alleles and genotype distribution
The allelic distribution of three TGF-β1 and eight IL-6 SNPs between OC patients and controls are summarized in Table 2. Our results showed only a negative significantly association between the minor allele of IL-6 rs1880242 and OC susceptibility (p = 0.0275; OR [95% CI] = 0.53 [0.29-0.97]).
Table 2
TGF-β1 and IL-6 SNPs allelic distribution in patients and controls
At genotypic level, comparing to C/C genotype of TGF-β1 rs1800469, the frequencies of C/T genotype were significantly lower in OC cases in under additive (p < 0.001; OR [95% CI] = 0.24 [0.15-0.58]) and dominant (p < 0.001, OR [95% CI] = 0.33 [0.15-0.74]) genetic models (Table 3). The genotype distributions of tested IL-6 variants, taking homozygous wild-type genotype as a reference (OR = 1.00), demonstrated that the combined genotypes of IL-6 rs1880242 [G/T+T/T] decreased the risk of OC (p = 0.021; OR [95% CI] = 0.38 [0.17-0.88]) (Table 4).
Table 3
TGF-β1 genotype distribution in cases and controls
Table 4
IL-6 genotype distribution in cases and controls
TGF-β1 and IL-6 haplotypes distribution
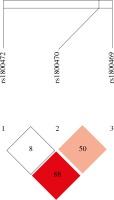
Table 5 presents the frequencies of three-locus TGF-β1 haplotypes in OC cases and controls, constructed based on the prevalence of individual SNPs and linkage disequilibrium (LD) between them. Of the 8 possible haplotypes, six were found to be common and were included in the analysis. Strong LD was found between TGF-β1 rs1800470, TGF-β1 rs1800472, and TGF-β1 rs1800469 (D’, 0.889; LOD, 4.28; r2, 0.093) (Fig. 1). Haplotype analysis showed that the distribution of TGF-β1 haplotypes was comparable between OC cases and control women, but not significant association was observed (Table 5).
Table 5
Distribution of 3-locus TGF-β1 haplotypes in ovarian cancer (OC) cases and controls
Fig. 1
Linkage disequilibrium (LD) plot and SNPs of TGF-β1. The plot was generated using Haploview program with D’ color scheme (D’ = 0, 0)
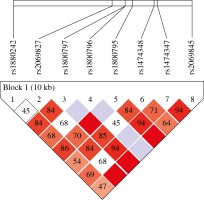
Furthermore, we evaluated the interaction between the tested IL-6 single nucleotide polymorphisms and by analyzing the haplotype distribution of 8-locus (IL-6 rs1880242, IL-6 rs2069827, IL-6 rs1800797, IL-6 rs1800796, IL-6 rs1800795, IL-6 rs1474348, IL-6 rs1474347, and IL-6 rs2069845) in OC cases and healthy controls by Haploview. Haplotype analysis demonstrated strong LD between IL-6 SNPs (Fig. 2).
Fig. 2
Linkage disequilibrium (LD) map of the six IL-6 SNPs genotyped using Haploview. The positions of tested SNPs are indicated above the Haploview output. The LD between specific pair of IL-6 SNPs is indicated by color scheme, which represents LD relationships, based on D’ values (normalized linkage disequilibrium measure or D) multiplied by 100. D’ is calculated as D divided by theoretical maximum for observed allele frequencies. Values approaching zero indicate absence of LD, and those approaching 100 indicate complete LD. The square-colored red represents varying degrees of LD < 1 and LOD (logarithm of odds) > 2 scores; darker shades indicating stronger LD
Results from Table 6 demonstrated development of GGAGGGGA and GGAGGGTA haplotypes and reduction in TGGGCCTA haplotype among OC cases than controls, thus conferring disease risk and protection to these IL-6 haplotypes, respectively. The distribution of other haplotypes was comparable between OC cases and control subjects.
Table 6
Distribution of 8-locus IL-6 haplotypes in OC cases and controls
Discussion
The exact mechanism causing the pathogenesis and increased susceptibility to OC remain unclear and controversial. It was previously suggested that chronic inflammatory changes are associated with the development and clinical course of OC [14]. In this regard, a central role of cytokines in cancer immunity was proposed, and it was originally suggested that altered expression of Th1-Th2 facilitates tumor growth through precipitation of initial state of tumor-specific and later, antigen non-specific immunosuppression (anergy) [15]. Recently, Th17 and Treg cells were described as key factors in regulating tumor growth. TGF-β1 is a multifunctional cytokine, which regulates the production of extracellular matrix, neovascularization, and immune response. Additionally, it promotes normal cell differentiation and motility, and is a crucial element in early stage of cancer [39, 40]. Owing to its action as angiogenic factor, TGF-β1 contributes to the pathology of advanced tumor stages, and is essential for the development, growth, invasion, and metastasis of cancer [41-43]. IL-6 has been previously known as hepatocyte-stimulating factor, cytotoxic T-cell differentiation factor, B-cell differentiation factor, B-cell stimulatory factor 2, hybridoma/plasmacytoma growth factor, monocyte granulocyte inducer type 2, and thrombopoietin, reflecting pleiotropism of this cytokine and its involvement in numerous biological functions [44-46].
It was established that IL-6 affects practically every organ, most notably the immune system. The physiological activity of IL-6 is very complex, including both pro-inflammatory and anti-inflammatory effects. This combination suggests that IL-6 may play a role in the control of immune system activation during various phases of epithelial ovarian cancer evolution [14, 47, 48].
In this study, we investigated the association of three TGF-β1 polymorphisms (rs1800469, rs1800470, and rs1800472) and eight IL-6 SNPs (rs1880242, rs2069827, rs1800797 rs1800796, rs1800795, rs1474348, rs1474347, and rs2069845) with the susceptibility to OC in Tunisians, the first to examine the possible association with OC in North African community.
Data obtained indicated that TGF-β1 rs1800469 heterozygosity was associated with a significant decrease in the risk of OC, thus assigning an OC protective nature for this variant. Previously, it was reported that carriage of the TGF-β1 rs1800469 minor allele, both in the heterozygous (C/T) and homozygous (T/T) states, did not affect the susceptibility to OC in Chinese population [49]. In contrast, a recent meta-analysis study that investigated the association between the TGF-β1 rs1800469 and cancers in different ethnic populations concluded that the TGF-β1 rs1800469 minor (T) allele is associated with a decreased risk of colorectal cancer, especially in Caucasians [50, 51]. These discrepancies may be reconciled by differences in ethnic origin between our Tunisian population and Asian women.
A possible association between the TGF-β1 rs1800469 and the risk of cancers yielded controversial results, in part due to the type of cancer and ethnic background of study participants [50-57]. Functionally, the TGF-β1 promoter variant rs1800469 (-509C/T) was associated with altered TGF-β1 plasma level and emphasized by the almost double level of circulating TGF-β1 in heterozygous (C/T) and homozygous (T/T) as compared to homozygous (C/C) genotype carriers [58]. Furthermore, it was previously shown that TGF-β1 is over-expressed in some human carcinomas, but not in all types of cancer, and that TGF- β1 mutations could influence its implication in cancer development [51, 58].
Heterozygosity of TGF-β1 rs1800469 and the presence of minor allele was associated with protection against OC in the study population. This was in agreement with a recent study, which documented association of TGF-β1 rs1800469 with a decreased risk of breast cancer in American women of African and European ancestry [54]. Neither TGF-β1 rs1800470 or TGF-β1 rs1800472 were markedly associated with OC in Tunisian women. This was the first report that examined the association between TGF-β1 variants and OC risk in Tunisia and worldwide. Previous studies examined the possible influence of genetic mutations in ovarian neoplasia and showed that TGF-β1 gene variants may influence (multistage) ovarian neoplasia by reducing epithelial cell responsiveness to TGF-β1 negative growth signals [23, 24].
For IL-6 gene polymorphisms, our results showed that only IL-6 rs1880242 was implicated in the occurrence of OC in Tunisia. Therefore, minor allele frequency (MAF) was considered as a risk factor, and the combined genotypes decreased the incidence of OC. The present study is the first to suggest a protective role of IL-6 rs1880242 in OC susceptibility. In fact, in Malaysian population, this SNP increased the risk to OC development [59]. These contradictory results for IL-6 rs1880242 and OC risk between Tunisians and Malaysians could be generated by the geographic and ethnical origin of populations, thus leading to the discordant findings between this polymorphism and OC occurrence. On the other hand, our data showed that IL-6 allele and genotype frequencies between OC cases with early tumor stages and healthy controls were comparable, but no significant association was detected between the seven IL-6 SNPs. In agreement with our results, previous study on Chinese population has reported a lack of association between IL-6 rs1800795 and OC risk [60].
It was established that IL-6 rs1800795 was associated with the plasma levels of protein [61]. However, some studies have investigated the possible association between this SNP, IL-6 plasma levels, and risk of cancers with controversial results [62-67].
In Tunisia, few case-control studies were conducted to evaluate the impact of IL-6 polymorphisms and cancers. A study by Snoussi et al. showed a significant association between SNPs in the promoter region IL-6 rs1800795 and IL-6 rs1800797 with breast cancer risk [68]. However, in our present study, we concluded that IL-6 rs1800795 and IL-6 rs1474348 were associated with cervical cancer susceptibility and evolution among Tunisian women [31].
On the other hand, our study suggests an association between IL-6 haplotypes (GGAGGGGA, GGAGGGTA and TGGGCCTA) and OC occurrence. To the best of our knowledge, no previous haplotypes association was detected between those eight IL-6 SNPs and OC susceptibility.
The present study has some strengths, including OC cases and control women of similar ethnicity, thus minimizing the possibility of admixture, and that only new OC cases prior to chemo- or radiotherapy were recruited. However, our studies had also some limitations, mostly due to the relatively limited sample size. Despite the incidence of OC in Tunisia, the rareness of this pathology lies in its quiet character responsible for a delay in diagnosis and therapeutic difficulty, especially in its extended forms. Therefore, the low sample size cannot lower the power of our study.
Conclusions
In the summary, this is the first study to show an association of TGF-β1 and IL-6 SNPs implicated in the inflammatory cascade with OC occurrence. We conclude that the TGF-β1 rs1800469, IL-6 rs1880242, and the IL-6 haplotype distribution may contribute to the occurrence of OC among Tunisian women. Subsequently, future studies will be performed to better understand the mechanism of TGF-β1 and IL-6 polymorphism involvement in changed plasma levels of these two proteins and their potential impact on OC.


