Introduction
Ovarian cancer is the seventh cause of death in females in the GLOBOCAN 2020 report. In 2020 this disease was diagnosed in 313 959 cases and resulted in the deaths of 207 252 women. The morbidity for ovarian cancer varies geographically, being the highest in Europe, and lowest in Africa [1]. Nowadays, many risk factors are known, including lifestyle – cigarette smoking, obesity, exposure to environmental agents; and reproductive factors – such as the number of pregnancies, infertility or hormone replacement therapy (HRT) [2, 3]. It seems that genetic factors play the most important role in developing ovarian cancer.
Recently, in the last decade, research has focused on finding new pathological pathways to the manifestation of tumors. MicroRNAs (miRNAs) are small, non-coding RNAs, containing approximately 21 to 23 nucleotides. They have been found not only in humans but also in plants, animals, and some viruses. They were first detected and reported in Caenorhabditis elegans in 1990 [4, 5]. Studies have proved their role in the development and progression of cancers, as well as other diseases, including asthma, rheumatic disorders, multiple sclerosis, depression, and many others [6–9]. These small particles play a role in the processes of degradation of RNA and the inhibition of messenger RNA translation, affecting transcription through connection with the 3′ untranslated region (3′UTR) of target messenger RNAs [10]. Dysregulation of miRNAs is characteristic for many types of tumors; moreover, the expression of specific miRNAs can be linked with specific tumors and stages of illness. Apart from the impact on tumor evolution, recent studies have demonstrated that miRNA can also adjust the tumor microenvironment (TME), allowing angiogenesis, immune invasion and metastasis [4].
MiRNAs originating from the 5′-end of human microRNA-196 hairpins play a role in controlling various biological functions such as cell cycle regulation, response to stress, growth, development, gene expression regulation (both transcriptionally and post-transcriptionally), protein metabolism, signaling, cellular homeostasis, chromosome arrangement and additional pathways. MiRNAs play a pivotal role not only in benign processes but also in malignant diseases. There are three loci that are members of the microRNA-196 family: miR-196a-1, miR-196a-2, and miR-196b. Nowadays, researchers worldwide are attempting to determine their role in tumorigenesis.
MiRNAs influence ovarian cancer progression by regulating the development of the cell cycle and apoptosis. An extensively researched single nucleotide polymorphism (SNP), rs11614913 [C>T], is situated within the hsa-mir-196a-2 locus, impacting the processing and expression of mature miRNAs derived from this locus [11]. The upregulation of miR-196a enhances ovarian cancer cell proliferation and reduces apoptosis by directly targeting the DEAD box protein RNA helicase 3 (DDX3) through regulation of the PTEN/PI3K/AKT pathway. DDX3 is found to be downregulated in ovarian cancer, and its decreased expression correlates with the progression of the disease [12]. In this particular case, miR-196-a also has many polymorphisms. The distribution of alleles and genotypes is different for distinct tumors.
MicroRNA-196a-1 and -2, through their involvement in cardiac development, may contribute to the development of ischemic heart disease in the future. Additionally, they serve as crucial indicators of comorbidity in type 2 diabetes – including cardiovascular system diseases and diabetic nephropathy [13]. The miR-196a-2 polymorphism shows significant associations with various types of cancer, including breast, lung, esophageal, gastric, and hepatocellular cancer. Detecting ovarian cancer early poses a challenge due to the absence of symptoms until the disease reaches advanced stages. Current methods for detecting ovarian cancer are not sufficient for diagnosing its early stages. Therefore, there is a need to focus on searching for additional biomarkers. The purpose of this study is to determine the relationship between the expression of microRNA 196-a along with their polymorphisms in ovarian cancer tissue and their impact on survival, clinical stage and chemosensitivity.
Material and methods
Patients
Healthy controls and patients with high-grade serous carcinoma (n = 50) were recruited for this prospective study. All participants signed written informed consent. The study received positive approval from the Bioethical Committee of the Polish Mother’s Memorial Hospital. Samples for testing were obtained from archival paraffin blocks obtained from intraoperative material. Additionally, the levels of Ca 125 and HE-4 were assessed in most cases. The study included patients who underwent surgery in the Department of Operative Gynaecology, Endoscopy and Gynecologic Oncology between 2010 and 2017. Clinical staging was determined according to the FIGO classification. The examination of tissue samples was carried out at the Department of Clinical Pathology within the Polish Mother’s Memorial Hospital – Research Institute, located in Lodz, Poland. Genetic tests focusing on microRNA-196a were conducted at the Research Laboratory CoreLab, which is part of the Medical University of Lodz, Poland.
The study was conducted among women aged between 30 and 83 years at any clinical stage of the disease (FIGO IA-IVB). The control group consisted of 10 participants. Ca 125 concentration values ranged from levels of 7 to 5000 units, while levels of HE-4 ranged from 17 to 1500 units. Calculated ROMA (the risk of ovarian malignancy algorithm) was from 10.2 to 99.5%. The body mass index (BMI) was also estimated: the lowest was 18 and the highest was 39. Among chronic diseases, hypertension was the most common (Table 1).
Table 1
Characteristics of patients
Genotyping of rs11614913 SNP in miR-196a
Genomic DNA was isolated from FFPE samples using the High Pure PCR Template Preparation Kit (Roche Diagnostics GmbH, Mannheim, Germany) following the manufacturer’s protocol. DNA was quantified using a Picodrop spectrophotometer (Picodrop Limited) and either used immediately for PCR reaction or stored at –20°C.
The miR-196a C/T single-nucleotide polymorphism rs11614913 was analyzed using the commercially available pre-made TaqMan SNP Genotyping Assay (Applied Biosystems, CA, USA) with the assay ID: C_31185852_10. PCR was performed using the GeneAmp PCR System 7900 (Applied Biosystems, CA, USA) in a 20 µl reaction volume containing 10 ng of DNA, 10 µl of TaqMan Universal PCR Master Mix, and 0.5 µl of 40× TaqMan SNP Genotyping Assay. The PCR thermal cycling conditions were: initial denaturing at 95°C for 10 minutes, 40 cycles of 92°C for 15 seconds and 60°C for 1 minute. Each 96-well plate included tested samples and three no-template controls (NTC). The end-point fluorescent intensities of each probe were monitored using the 7900HT Fast Real-Time PCR System (Applied Biosystems, CA, USA). The genotypes were determined automatically and visually verified using Sequence Detection System 2.3 Software.
miRNA expression analysis
Total RNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissues using the High Pure miRNA Isolation Kit (Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturer’s instructions. Briefly, FFPE samples were deparaffinized with 800 µl of 100% xylene for 5 minutes, washed in 1 ml of 100% ethanol, and dried at 55°C for 10 minutes. 100 µl of paraffin tissue lysis buffer, 16 µl of 10% SDS, and 40 µl of proteinase K working solution were added to the dried tissue and incubated at 55°C overnight. Following tissue lysis, RNA was purified on columns according to the manufacturer’s protocol. 325 µl of binding buffer and 325 µl of binding enhancer were added, and the mixture was applied to a high pure filter tube. Then columns were centrifuged for 30 seconds at 13,000 × g and washed twice with 500 µl and 300 µl of wash buffer. An additional centrifugation step was performed to dry the filter completely. RNA was eluted with 50 µl of elution buffer. RNA concentration and quality (260/280 optical density ratios) were determined using a Picodrop spectrophotometer (Picodrop Limited, Hinxton, UK). The purified total RNA was immediately used for cDNA synthesis or stored at –80°C until use.
The reverse transcription of 10 µg of the total RNA was carried out using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, CA, USA), following the manufacturer’s instructions. TaqMan PCR assays were performed on the 7900HT Fast Real-Time PCR System (Applied Biosystems, CA, USA). miRNA quantification was done using standard TaqMan MicroRNA Assays (Applied Biosystems, CA, USA): hsa-miR196a (Assay ID: 241070 mat), and RNU6B (Assay ID: 001093) as a control. The 20 µl of qPCR reaction mixture included 1.33 µl of RT product, 10 µl of TaqMan Universal PCR Master Mix and 1 µl of TaqMan miRNA Assay (20×). The reactions were incubated in a 96-well plate at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 600°C for 1 minute. All reactions were run in duplicate and analyzed using Sequence Detection System 2.3 Software. Expression values were calculated using Sequence Detection System 2.3 Software and Data Assist Software (Applied Biosystems, CA, USA).
Statistical analysis
The relationship between miR-196a expression levels and patient survival rates was assessed using Kaplan-Meier survival curves. Group differences were assessed using the Mann-Whitney U test. Spearman’s rank correlation coefficient was employed to analyze the statistical dependence between two variables. Multivariate analysis was conducted to estimate correlations involving three or more variables. A significance level of p < 0.05 was used to denote statistical significance.
Results
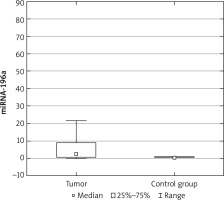
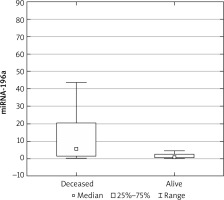
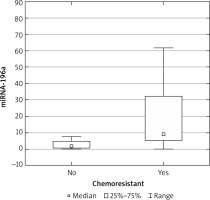
The expression results in the control groups were consistently convergent. The examinations conducted on specimens revealed elevated miRNA 196a expression within the cohort of patients diagnosed with ovarian cancer, as compared with the control group (p < 0.05) (Fig. 1). Moreover, elevated expression was observed among patients with more advanced FIGO stages (III, IV) – a result of borderline statistical significance (p = 0.05). Increased miRNA expression was also associated with significantly higher mortality (Fig. 2) and chemoresistance (p < 0.05) (Fig. 3).
We investigated the potential correlation between concentrations of Ca 125 and HE-4 and miRNA expression; however, no such association was detected. An analysis of the frequency of relevant SNPs (C/C, C/T, T/T) was also conducted between the control and study groups. In the study group, the most common genotype was C/C, followed by C/T and T/T. In the control group, the obtained results were partially opposite: the T/T genotype was the most common in this group. However, statistically significant correlations between the frequency of mentioned genotypes and the occurrence of malignancy were not observed (Table 2).
Discussion
In the introduction, we presented an extensive literature review regarding the significance of microRNAs. We know that these molecules play a role in key processes related to tumorigenesis and can also serve as prognostic factors. In the available literature, scientists primarily focus on the significance of the microRNAs we are studying in epithelial-derived cancers, mainly those of the gastrointestinal and respiratory tracts. Among malignant tumors of the reproductive system, research is ongoing regarding the importance of their expression in ovarian or endometrial cancer. In our study, we conducted an analysis of miR-196a expression among patients diagnosed with ovarian cancer compared to a control group consisting of healthy individuals. The aim of the study was to demonstrate the relationship between microRNA expression and its impact on survival and response to chemotherapy, correlated with the clinical data of the patients participating in the study. Up to the time of preparing this publication, only a few articles evaluating such an association can be found in the available medical databases.
Fan et al. found a correlation between miR-196a expression and tumor stage, tumor size, and lymph node metastasis. The expression level of miR-196a was elevated in epithelial ovarian cancer patients with advanced-stage tumors (p < 0.001). Furthermore, high miR-196a expression was more commonly observed in patients with larger tumors (p = 0.020). No significant correlations were found between miR-196a expression and other clinicopathologic features, such as age, histological type, or grade. Patients with higher miR-196a expression exhibited shorter both overall (p < 0.001) and recurrence-free survival (p = 0.003) compared to those with lower miR-196a expression [14]. The results obtained in that study are similar to ours.
Song et al. conducted a study among 475 patients with epithelial ovarian cancer and 474 healthy individuals, seeking to establish correlations between the frequency of specific polymorphisms in both study groups. The results showed a notably higher prevalence of CT and CC genotypes among the patients compared to the control group (51.6% vs. 47.1%, p = 0.063 and 25.2% vs. 20.0%, p = 0.018). Further grouping of the TT and CT genotypes in the recessive genetic model revealed a significantly elevated risk of ovarian cancer in CC genotype carriers compared to wild-type TT homozygotes and CT heterozygotes (OR = 1.36; 95% CI: 1.04–2.17; p = 0.023) [15]. The C/C genotype was also the most common in our study group; however, no association was found between the frequency of a given genotype and the risk of developing a malignant tumor.
Liu et al. conducted a systematic review and meta-analysis aiming to comprehensively analyze and summarize the association between non-coding RNA polymorphisms and the risk of ovarian cancer. Significant associations between miR-196a rs11614913 and ovarian cancer risk were observed in three genetic models: T vs. C (allele model): OR = 0.70, 95% CI: 0.59–0.82; TT + TC vs. CC (dominant model): OR = 1.62, 95% CI: 1.18–2.24; and TT vs. TC + CC (recessive model): OR = 0.70, 95% CI: 0.57–0.87. The T genotype may decrease susceptibility to ovarian cancer. However, no association was found in the following two genetic models: TC vs. CC (heterozygote model): OR = 1.28, 95% CI: 0.98–1.67, and TT vs. CC (homozygous model): OR = 2.36, 95% CI: 0.86–6.44. The results obtained from the mentioned studies were not corroborated in our study; however, this discrepancy may stem from the smaller sample size in our research [16].
Conclusions
In summary, this study highlights the critical role of microRNA-196a in the progression of ovarian cancer, with significant implications for patient survival and response to chemotherapy. Our findings indicate that elevated expression of miR-196a is associated with advanced stages of ovarian cancer, increased mortality, and higher chemoresistance. These results underscore the potential of miR-196a as a biomarker for prognosis and therapeutic targeting in ovarian cancer.
Despite the clear association between miR-196a expression and ovarian cancer advancement, the study did not establish a significant correlation between the single nucleotide polymorphism (SNP) rs11614913 within miR-196a-2 and the incidence of ovarian cancer. This discrepancy with previous studies might be attri-buted to the limited sample size in our research.
The clinical utility of miR-196a as a biomarker warrants further investigation, particularly in larger, more diverse cohorts. Understanding the molecular mechanisms by which miR-196a influences tumor biology could pave the way for novel therapeutic strategies aimed at modulating its expression. Future research should also explore the integration of miR-196a analysis with other diagnostic markers to enhance early detection and personalized treatment approaches for ovarian cancer.
Ultimately, this study contributes to the growing body of evidence supporting the significance of miRNAs in cancer biology and highlights the need for ongoing research to fully elucidate their potential in clinical applications.